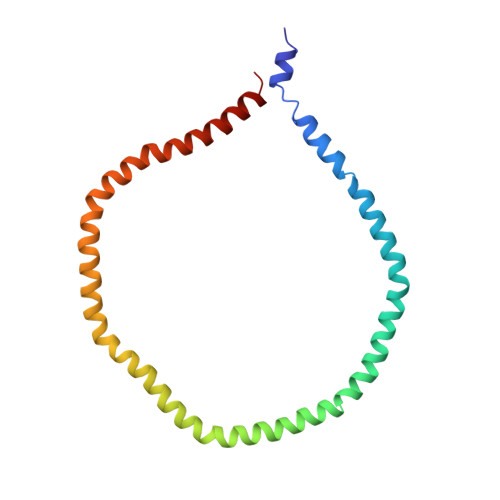
Solution structure of discoidal high-density lipoprotein particles with a shortened apolipoprotein A-I.
Bibow, S., Polyhach, Y., Eichmann, C., Chi, C.N., Kowal, J., Albiez, S., McLeod, R.A., Stahlberg, H., Jeschke, G., Guntert, P., Riek, R.(2017) Nat Struct Mol Biol 24: 187-193
- PubMed: 28024148
- DOI: https://doi.org/10.1038/nsmb.3345
- Primary Citation of Related Structures:
2N5E - PubMed Abstract:
High-density lipoprotein (HDL) particles are cholesterol and lipid transport containers. Mature HDL particles destined for the liver develop through the formation of intermediate discoidal HDL particles, which are the primary acceptors for cholesterol. Here we present the three-dimensional structure of reconstituted discoidal HDL (rdHDL) particles, using a shortened construct of human apolipoprotein A-I, determined from a combination of nuclear magnetic resonance (NMR), electron paramagnetic resonance (EPR) and transmission electron microscopy (TEM) data. The rdHDL particles feature a protein double belt surrounding a lipid bilayer patch in an antiparallel fashion. The integrity of this structure is maintained by up to 28 salt bridges and a zipper-like pattern of cation-π interactions between helices 4 and 6. To accommodate a hydrophobic interior, a gross 'right-to-right' rotation of the helices after lipidation is necessary. The structure reflects the complexity required for a shuttling container to hold a fluid lipid or cholesterol interior at a protein:lipid ratio of 1:50.
- Laboratory of Physical Chemistry, ETH Zurich, Zurich, Switzerland.
Organizational Affiliation: