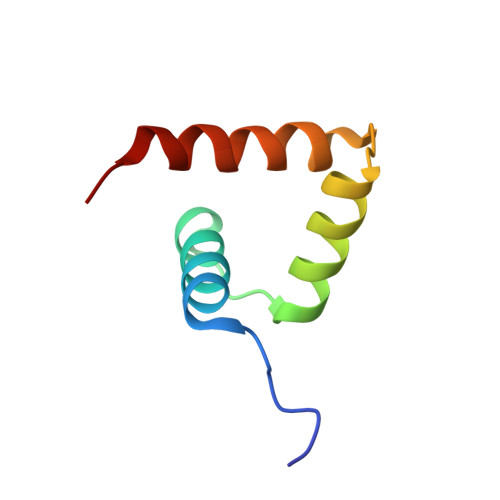
Solution structure of the porcine sapovirus VPg core reveals a stable three-helical bundle with a conserved surface patch.
Hwang, H.J., Min, H.J., Yun, H., Pelton, J.G., Wemmer, D.E., Cho, K.O., Kim, J.S., Lee, C.W.(2015) Biochem Biophys Res Commun 459: 610-616
- PubMed: 25753201
- DOI: https://doi.org/10.1016/j.bbrc.2015.02.156
- Primary Citation of Related Structures:
2MXD - PubMed Abstract:
Viral protein genome-linked (VPg) proteins play a critical role in the life cycle of vertebrate and plant positive-sense RNA viruses by acting as a protein primer for genome replication and as a protein cap for translation initiation. Here we report the solution structure of the porcine sapovirus VPg core (VPg(C)) determined by multi-dimensional NMR spectroscopy. The structure of VPg(C) is composed of three α-helices stabilized by several conserved hydrophobic residues that form a helical bundle core similar to that of feline calicivirus VPg. The putative nucleotide acceptor Tyr956 within the first helix of the core is completely exposed to solvent accessible surface to facilitate nucleotidylation by viral RNA polymerase. Comparison of VPg structures suggests that the surface for nucleotidylation site is highly conserved among the Caliciviridae family, whereas the backbone core structures are different. These structural features suggest that caliciviruses share common mechanisms of VPg-dependent viral replication and translation.
- Department of Chemistry, Chonnam National University, Gwangju 500-757, Republic of Korea.
Organizational Affiliation: