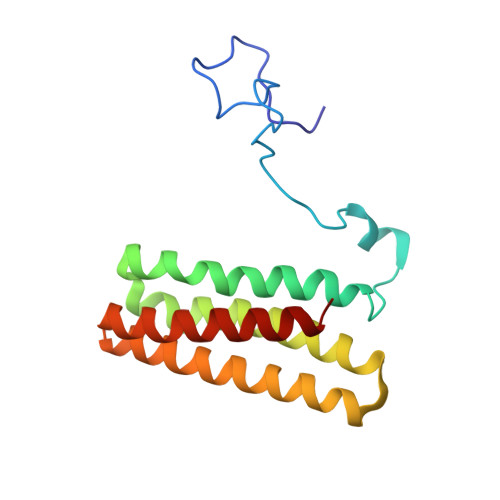
Solution NMR and molecular dynamics reveal a persistent alpha helix within the dynamic region of PsbQ from photosystem II of higher plants.
Rathner, P., Rathner, A., Hornicakova, M., Wohlschlager, C., Chandra, K., Kohoutova, J., Ettrich, R., Wimmer, R., Muller, N.(2015) Proteins 83: 1677-1686
- PubMed: 26138376
- DOI: https://doi.org/10.1002/prot.24853
- Primary Citation of Related Structures:
2MWQ - PubMed Abstract:
The extrinsic proteins of photosystem II of higher plants and green algae PsbO, PsbP, PsbQ, and PsbR are essential for stable oxygen production in the oxygen evolving center. In the available X-ray crystallographic structure of higher plant PsbQ residues S14-Y33 are missing. Building on the backbone NMR assignment of PsbQ, which includes this "missing link", we report the extended resonance assignment including side chain atoms. Based on nuclear Overhauser effect spectra a high resolution solution structure of PsbQ with a backbone RMSD of 0.81 Å was obtained from torsion angle dynamics. Within the N-terminal residues 1-45 the solution structure deviates significantly from the X-ray crystallographic one, while the four-helix bundle core found previously is confirmed. A short α-helix is observed in the solution structure at the location where a β-strand had been proposed in the earlier crystallographic study. NMR relaxation data and unrestrained molecular dynamics simulations corroborate that the N-terminal region behaves as a flexible tail with a persistent short local helical secondary structure, while no indications of forming a β-strand are found.
- Institute of Organic Chemistry, Johannes Kepler University Linz, Linz, 4040, Austria.
Organizational Affiliation: