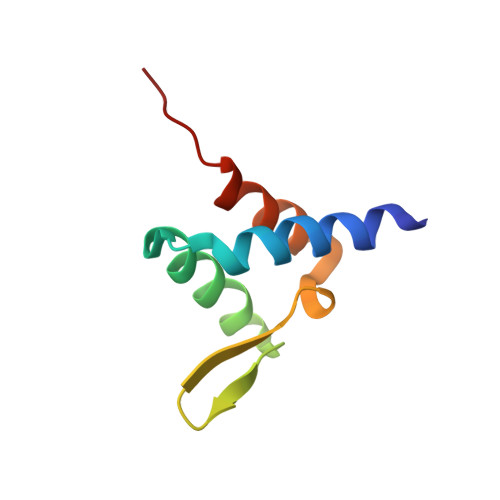
Degree of Recruitment of DOT1L to MLL-AF9 Defines Level of H3K79 Di- and Tri-methylation on Target Genes and Transformation Potential.
Kuntimaddi, A., Achille, N.J., Thorpe, J., Lokken, A.A., Singh, R., Hemenway, C.S., Adli, M., Zeleznik-Le, N.J., Bushweller, J.H.(2015) Cell Rep 11: 808-820
- PubMed: 25921540
- DOI: https://doi.org/10.1016/j.celrep.2015.04.004
- Primary Citation of Related Structures:
2MV7 - PubMed Abstract:
The MLL gene is a common target of chromosomal translocations found in human leukemia. MLL-fusion leukemia has a consistently poor outcome. One of the most common translocation partners is AF9 (MLLT3). MLL-AF9 recruits DOT1L, a histone 3 lysine 79 methyltransferase (H3K79me1/me2/me3), leading to aberrant gene transcription. We show that DOT1L has three AF9 binding sites and present the nuclear magnetic resonance (NMR) solution structure of a DOT1L-AF9 complex. We generate structure-guided point mutations and find that they have graded effects on recruitment of DOT1L to MLL-AF9. Chromatin immunoprecipitation sequencing (ChIP-seq) analyses of H3K79me2 and H3K79me3 show that graded reduction of the DOT1L interaction with MLL-AF9 results in differential loss of H3K79me2 and me3 at MLL-AF9 target genes. Furthermore, the degree of DOT1L recruitment is linked to the level of MLL-AF9 hematopoietic transformation.
- Department of Molecular Physiology and Biological Physics, University of Virginia, Charlottesville, VA 22908, USA.
Organizational Affiliation: