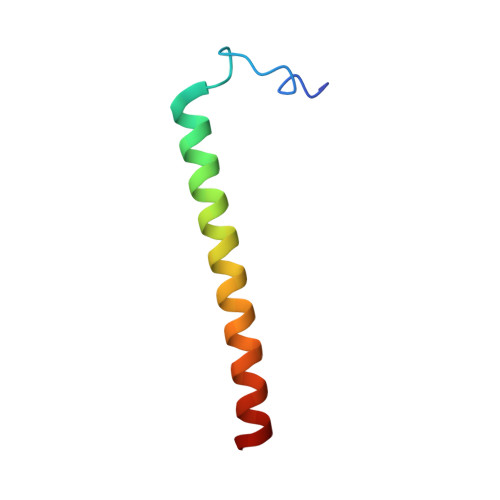
Structural insight into the transmembrane domain and the juxtamembrane region of the erythropoietin receptor in micelles.
Li, Q., Wong, Y.L., Huang, Q., Kang, C.(2014) Biophys J 107: 2325-2336
- PubMed: 25418301
- DOI: https://doi.org/10.1016/j.bpj.2014.10.013
- Primary Citation of Related Structures:
2MV6 - PubMed Abstract:
Erythropoietin receptor (EpoR) dimerization is an important step in erythrocyte formation. Its transmembrane domain (TMD) and juxtamembrane (JM) region are essential for signal transduction across the membrane. A construct compassing residues S212-P259 and containing the TMD and JM region of the human EpoR was purified and reconstituted in detergent micelles. The solution structure of the construct was determined in dodecylphosphocholine (DPC) micelles by solution NMR spectroscopy. Structural and dynamic studies demonstrated that the TMD and JM region are an ?-helix in DPC micelles, whereas residues S212-D224 at the N-terminus of the construct are not structured. The JM region is a helix that contains a hydrophobic patch formed by conserved hydrophobic residues (L253, I257, and W258). Nuclear Overhauser effect analysis, fluorescence spectroscopy, and paramagnetic relaxation enhancement experiments suggested that the JM region is exposed to the solvent. The structures of the TMD and JM region of the mouse EpoR were similar to those of the human EpoR.
- Institute of Chemical & Engineering Sciences, Technology and Research (A(?)STAR), Singapore.
Organizational Affiliation: