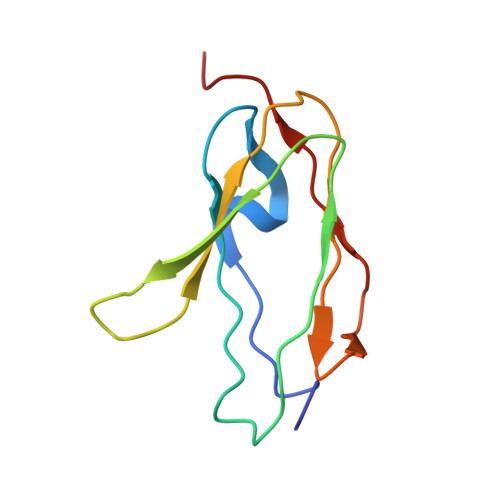
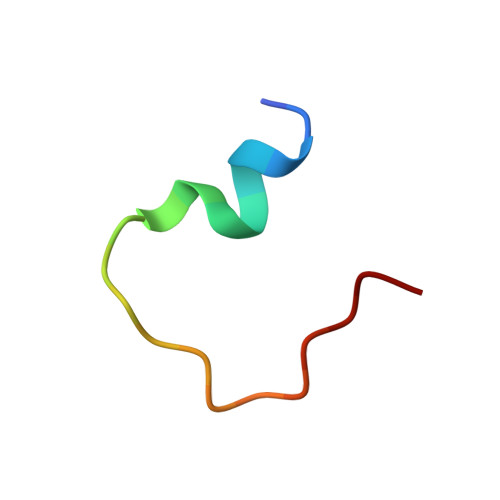
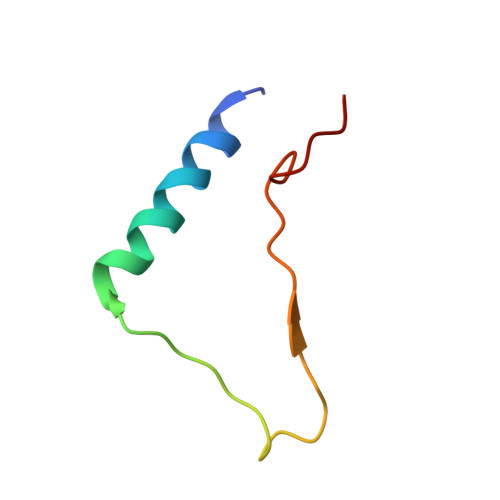
Structural mechanism of integrin inactivation by filamin.
Liu, J., Das, M., Yang, J., Ithychanda, S.S., Yakubenko, V.P., Plow, E.F., Qin, J.(2015) Nat Struct Mol Biol 22: 383-389
- PubMed: 25849143
- DOI: https://doi.org/10.1038/nsmb.2999
- Primary Citation of Related Structures:
2MTP - PubMed Abstract:
Activation of heterodimeric (αβ) integrin is crucial for regulating cell adhesion. Binding of talin to the cytoplasmic face of integrin activates the receptor, but how integrin is maintained in a resting state to counterbalance its activation has remained obscure. Here, we report the structure of the cytoplasmic domain of human integrin αIIbβ3 bound to its inhibitor, the immunoglobin repeat 21 of filamin A (FLNa-Ig21). The structure reveals an unexpected ternary complex in which FLNa-Ig21 not only binds to the C terminus of the integrin β3 cytoplasmic tail (CT), as previously predicted, but also engages N-terminal helices of αIIb and β3 CTs to stabilize an inter-CT clasp that helps restrain the integrin in a resting state. Combined with functional data, the structure reveals a new mechanism of filamin-mediated retention of inactive integrin, suggesting a new framework for understanding regulation of integrin activation and adhesion.
- Department of Molecular Cardiology, Lerner Research Institute, Cleveland Clinic, Cleveland, Ohio, USA.
Organizational Affiliation: