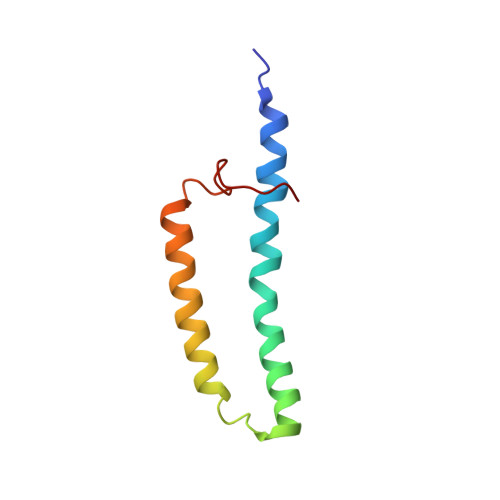
Structure of the membrane protein MerF, a bacterial mercury transporter, improved by the inclusion of chemical shift anisotropy constraints.
Tian, Y., Lu, G.J., Marassi, F.M., Opella, S.J.(2014) J Biomol NMR 60: 67-71
- PubMed: 25103921
- DOI: https://doi.org/10.1007/s10858-014-9852-0
- Primary Citation of Related Structures:
2MOZ - PubMed Abstract:
MerF is a mercury transport membrane protein from the bacterial mercury detoxification system. By performing a solid-state INEPT experiment and measuring chemical shift anisotropy frequencies in aligned samples, we are able to improve on the accuracy and precision of the initial structure that we presented. MerF has four N-terminal and eleven C-terminal residues that are mobile and unstructured in phospholipid bilayers. The structure presented here has average pairwise RMSDs of 1.78 Å for heavy atoms and 0.92 Å for backbone atoms.
- Department of Chemistry and Biochemistry, University of California San Diego, 9500 Gilman Drive, La Jolla, CA, 92093-0307, USA.
Organizational Affiliation: