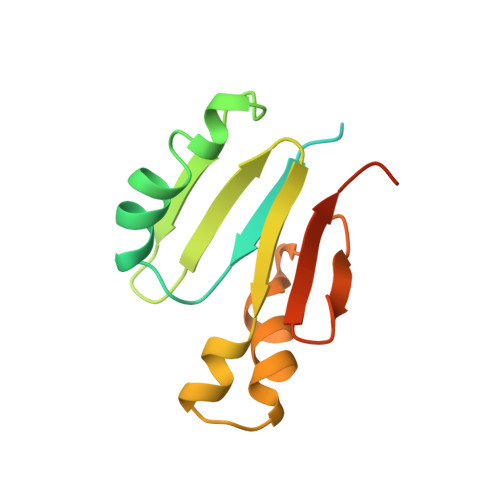
Solution structure of component B from methane monooxygenase derived through heteronuclear NMR and molecular modeling.
Chang, S.L., Wallar, B.J., Lipscomb, J.D., Mayo, K.H.(1999) Biochemistry 38: 5799-5812
- PubMed: 10231531
- DOI: https://doi.org/10.1021/bi982992f
- Primary Citation of Related Structures:
2MOB - PubMed Abstract:
Methane monooxygenase (MMO) is a nonheme iron-containing enzyme which consists of three protein components: a hydroxylase (MMOH), an NADH-linked reductase (MMOR), and a small "B" component (MMOB) which plays a regulatory role. Here, 1H, 13C, 15N heteronuclear 2D and 3D NMR spectroscopy has been used to derive the solution structure of the 138 amino acid MMOB protein in the monomer state. Pulse field gradient NMR self-diffusion measurements indicate predominant formation of dimers at 1 mM MMOB and monomers at or below 0.2 mM. MMOB is active as a monomer. Aggregate exchange broadening and limited solubility dictated that multidimensional heteronuclear NMR experiments had to be performed at a protein concentration of 0.2 mM. Using 1340 experimental constraints (1182 NOEs, 98 dihedrals, and 60 hydrogen bonding) within the well-folded part of the protein (residues 36-126), MMOB structural modeling produced a well-defined, compact alpha/beta fold which consists of three alpha-helices and six antiparallel beta-strands arranged in two domains: a betaalphabetabeta and a betaalphaalphabetabeta. Excluding the ill-defined N- and C-terminal segments (residues 1-35 and 127-138), RMS deviations are 1.1 A for backbone atoms and 1.6 A for all non-hydrogen atoms. Compared to the lower resolution NMR structure for the homologous protein P2 from the Pseudomonas sp. CF600 phenol hydroxylase system (RMSD = 2.48 A for backbone atoms) (Qian, H., Edlund, U., Powlowski, J., Shingler, V., and Sethson, I. (1997) Biochemistry, 36, 495-504), that of MMOB reveals a considerably more compact protein. In particular, MMOB lacks the large "doughnut" shaped cavity reported for the P2 protein. This difference may result from the limited number of long-range NOEs that were available for use in the modeling of the P2 structure. This NMR-derived structure of MMOB, therefore, presents the first high-resolution structure of a small protein effector of a nonheme oxygenase system.
- Department of Biochemistry, Molecular Biology & Biophysics, Center for Metals in Biocatalysis, University of Minnesota, Minneapolis 55455, USA.
Organizational Affiliation: