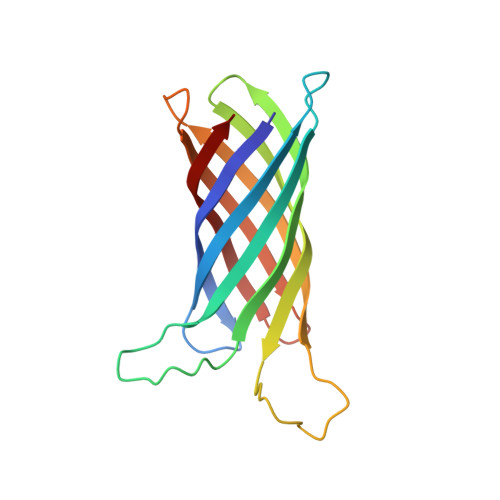
Measuring membrane protein bond orientations in nanodiscs via residual dipolar couplings.
Bibow, S., Carneiro, M.G., Sabo, T.M., Schwiegk, C., Becker, S., Riek, R., Lee, D.(2014) Protein Sci 23: 851-856
- PubMed: 24752984
- DOI: https://doi.org/10.1002/pro.2482
- Primary Citation of Related Structures:
2MNH - PubMed Abstract:
Membrane proteins are involved in numerous vital biological processes. To understand membrane protein functionality, accurate structural information is required. Usually, structure determination and dynamics of membrane proteins are studied in micelles using either solution state NMR or X-ray crystallography. Even though invaluable information has been obtained by this approach, micelles are known to be far from ideal mimics of biological membranes often causing the loss or decrease of membrane protein activity. Recently, nanodiscs, which are composed of a lipid bilayer surrounded by apolipoproteins, have been introduced as a more physiological alternative than micelles for NMR investigations on membrane proteins. Here, we show that membrane protein bond orientations in nanodiscs can be obtained by measuring residual dipolar couplings (RDCs) with the outer membrane protein OmpX embedded in nanodiscs using Pf1 phage as an alignment medium. The presented collection of membrane protein RDCs in nanodiscs represents an important step toward more comprehensive structural and dynamical NMR-based investigations of membrane proteins in a natural bilayer environment.
- Laboratory for Physical Chemistry, ETH Zürich, CH-8093, Zürich, Switzerland.
Organizational Affiliation: