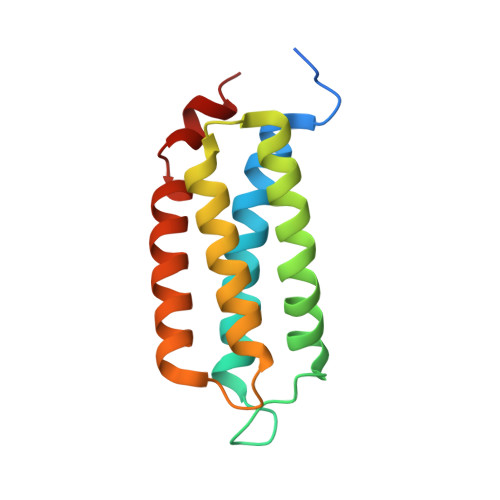
An oxygen-sensitive toxin-antitoxin system.
Marimon, O., Teixeira, J.M., Cordeiro, T.N., Soo, V.W., Wood, T.L., Mayzel, M., Amata, I., Garcia, J., Morera, A., Gay, M., Vilaseca, M., Orekhov, V.Y., Wood, T.K., Pons, M.(2016) Nat Commun 7: 13634-13634
- PubMed: 27929062
- DOI: https://doi.org/10.1038/ncomms13634
- Primary Citation of Related Structures:
2MN2 - PubMed Abstract:
The Hha and TomB proteins from Escherichia coli form an oxygen-dependent toxin-antitoxin (TA) system. Here we show that YmoB, the Yersinia orthologue of TomB, and its single cysteine variant [C117S]YmoB can replace TomB as antitoxins in E. coli. In contrast to other TA systems, [C117S]YmoB transiently interacts with Hha (rather than forming a stable complex) and enhances the spontaneous oxidation of the Hha conserved cysteine residue to a -SO x H-containing species (sulfenic, sulfinic or sulfonic acid), which destabilizes the toxin. The nuclear magnetic resonance structure of [C117S]YmoB and the homology model of TomB show that the two proteins form a four-helix bundle with a conserved buried cysteine connected to the exterior by a channel with a diameter comparable to that of an oxygen molecule. The Hha interaction site is located on the opposite side of the helix bundle.
- Biomolecular NMR Laboratory, Organic Chemistry Section, Inorganic and Organic Chemistry Department, University of Barcelona, Baldiri Reixac 10-12, Barcelona 08028, Spain.
Organizational Affiliation: