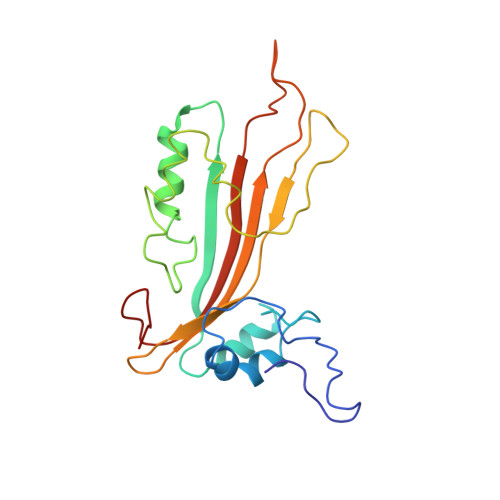
New Cyt-like delta-endotoxins from Dickeya dadantii: structure and aphicidal activity.
Loth, K., Costechareyre, D., Effantin, G., Rahbe, Y., Condemine, G., Landon, C., da Silva, P.(2015) Sci Rep 5: 8791-8791
- PubMed: 25740111
- DOI: https://doi.org/10.1038/srep08791
- Primary Citation of Related Structures:
2MLW - PubMed Abstract:
In the track of new biopesticides, four genes namely cytA, cytB, cytC and cytD encoding proteins homologous to Bacillus thuringiensis (Bt) Cyt toxins have been identified in the plant pathogenic bacteria Dickeya dadantii genome. Here we show that three Cyt-like δ-endotoxins from D. dadantii (CytA, CytB and CytC) are toxic to the pathogen of the pea aphid Acyrthosiphon pisum in terms of both mortality and growth rate. The phylogenetic analysis of the comprehensive set of Cyt toxins available in genomic databases shows that the whole family is of limited taxonomic occurrence, though in quite diverse microbial taxa. From a structure-function perspective the 3D structure of CytC and its backbone dynamics in solution have been determined by NMR. CytC adopts a cytolysin fold, structurally classified as a Cyt2-like protein. Moreover, the identification of a putative lipid binding pocket in CytC structure, which has been probably maintained in most members of the Cyt-toxin family, could support the importance of this lipid binding cavity for the mechanism of action of the whole family. This integrative approach provided significant insights into the evolutionary and functional history of D. dadantii Cyt toxins, which appears to be interesting leads for biopesticides.
- Centre de Biophysique Moléculaire, CNRS UPR 4301, Université d'Orléans, Orléans, F-45071, France.
Organizational Affiliation: