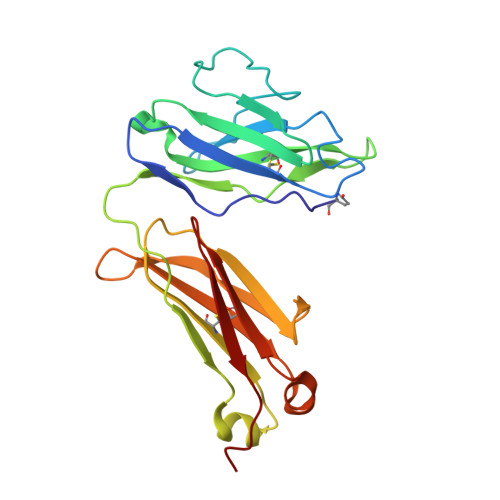
Three-dimensional structure of a light chain dimer crystallized in water. Conformational flexibility of a molecule in two crystal forms.
Ely, K.R., Herron, J.N., Harker, M., Edmundson, A.B.(1989) J Mol Biology 210: 601-615
- PubMed: 2515285
- DOI: https://doi.org/10.1016/0022-2836(89)90135-6
- Primary Citation of Related Structures:
1DCL, 2MCG, 3MCG - PubMed Abstract:
The three-dimensional structure of an immunoglobulin light chain dimer (Mcg) crystallized in deionized water (orthorhombic form) was determined at 2.0 A resolution by phase extension and crystallographic refinement. This structure was refined side-by-side with that of the same molecule crystallized in ammonium sulfate (trigonal form). The dimer adopted markedly different structures in the two solvents. "Elbow bend" angles between pseudo 2-fold axes of rotation relating pairs of "variable" (V) and "constant" (C) domains were found to be 132 degrees in the orthorhombic form and 115 degrees in the trigonal form. Modes of association of the V domains and, to a lesser extent, the pairing interactions of the C domains were different in the two structures. Alterations in the V domain pairing were reflected in the shapes of the binding regions and in the orientations of the side-chains lining the walls of the binding sites. In the trigonal form, for instance, the V domain interface was compartmentalized into a main binding cavity and a deep pocket, whereas these spaces were continuous in the orthorhombic structure. Patterns of ordered water molecules were quite distinct in the two crystal types. In some cases, the solvent structures could be correlated with conformational changes in the proteins. For example, close contacts between V and C domains of monomer 1 of the trigonal form were not retained in orthorhombic crystals. Ordered water molecules filled the space created when the two domains moved apart.
- Department of Biology, University of Utah, Salt Lake City 84112.
Organizational Affiliation: