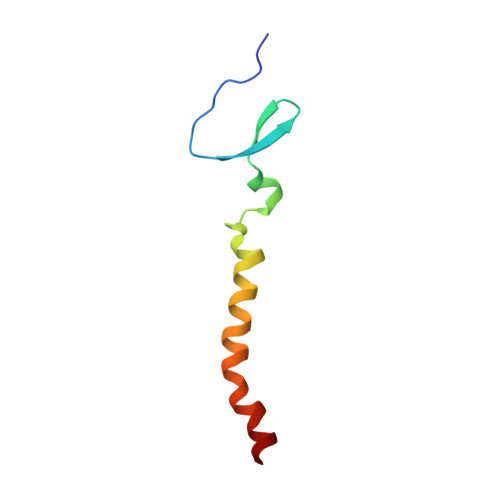
Structure of an HIV-1-neutralizing antibody target, the lipid-bound gp41 envelope membrane proximal region trimer.
Reardon, P.N., Sage, H., Dennison, S.M., Martin, J.W., Donald, B.R., Alam, S.M., Haynes, B.F., Spicer, L.D.(2014) Proc Natl Acad Sci U S A 111: 1391-1396
- PubMed: 24474763
- DOI: https://doi.org/10.1073/pnas.1309842111
- Primary Citation of Related Structures:
2LP7, 2M7W - PubMed Abstract:
The membrane proximal external region (MPER) of HIV-1 glycoprotein (gp) 41 is involved in viral-host cell membrane fusion. It contains short amino acid sequences that are binding sites for the HIV-1 broadly neutralizing antibodies 2F5, 4E10, and 10E8, making these binding sites important targets for HIV-1 vaccine development. We report a high-resolution structure of a designed MPER trimer assembled on a detergent micelle. The NMR solution structure of this trimeric domain, designated gp41-M-MAT, shows that the three MPER peptides each adopt symmetric α-helical conformations exposing the amino acid side chains of the antibody binding sites. The helices are closely associated at their N termini, bend between the 2F5 and 4E10 epitopes, and gradually separate toward the C termini, where they associate with the membrane. The mAbs 2F5 and 4E10 bind gp41-M-MAT with nanomolar affinities, consistent with the substantial exposure of their respective epitopes in the trimer structure. The traditional structure determination of gp41-M-MAT using the Xplor-NIH protocol was validated by independently determining the structure using the DISCO sparse-data protocol, which exploits geometric arrangement algorithms that guarantee to compute all structures and assignments that satisfy the data.
- Department of Biochemistry, Duke Human Vaccine Institute, Duke Department of Medicine, and Department of Radiology, Duke University School of Medicine, Durham, NC 27710.
Organizational Affiliation: