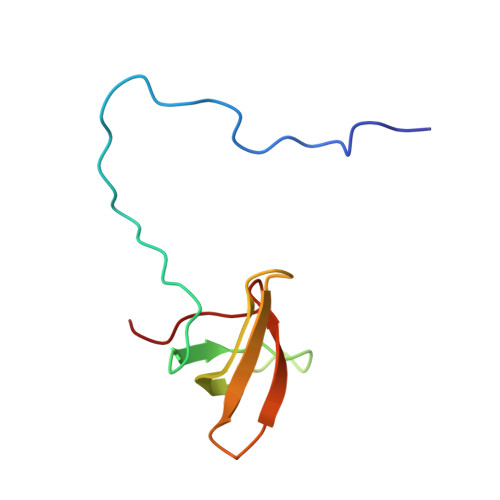
An H3K36 Methylation-Engaging Tudor Motif of Polycomb-like Proteins Mediates PRC2 Complex Targeting.
Cai, L., Rothbart, S.B., Lu, R., Xu, B., Chen, W.Y., Tripathy, A., Rockowitz, S., Zheng, D., Patel, D.J., Allis, C.D., Strahl, B.D., Song, J., Wang, G.G.(2013) Mol Cell 49: 571-582
- PubMed: 23273982
- DOI: https://doi.org/10.1016/j.molcel.2012.11.026
- Primary Citation of Related Structures:
2M0O - PubMed Abstract:
Polycomb repressive complex 2 (PRC2) regulates pluripotency, differentiation, and tumorigenesis through catalysis of histone H3 lysine 27 trimethylation (H3K27me3) on chromatin. However, the mechanisms that underlie PRC2 recruitment and spreading on chromatin remain unclear. Here we report that histone H3 lysine 36 trimethylation (H3K36me3) binding activity is harbored in the Tudor motifs of PRC2-associated polycomb-like (PCL) proteins PHF1/PCL1 and PHF19/PCL3. Ectopically expressed PHF1 induced Tudor-dependent stabilization of PRC2 complexes on bulk chromatin and mediated spreading of PRC2 and H3K27me3 into H3K36me3-containing chromatin regions. In murine pluripotent stem cells, we identified coexistence of H3K36me3, H3K27me3, and PHF19/PCL3 at a subset of poised developmental genes and demonstrated that PHF19/PCL3 Tudor function is required for optimal H3K27me3 and repression of these loci. Collectively, our data suggest that PCL recognition of H3K36me3 promotes intrusion of PRC2 complexes into active chromatin regions to promote gene silencing and modulate the chromatin landscape during development.
- UNC Lineberger Comprehensive Cancer Center, University of North Carolina at Chapel Hill School of Medicine, Chapel Hill, NC 27599, USA.
Organizational Affiliation: