Genomic and structural characterization of Kunitz-type peptide LmKTT-1a highlights diversity and evolution of scorpion potassium channel toxins.
Chen, Z., Luo, F., Feng, J., Yang, W., Zeng, D., Zhao, R., Cao, Z., Liu, M., Li, W., Jiang, L., Wu, Y.(2013) PLoS One 8: e60201-e60201
- PubMed: 23573241
- DOI: https://doi.org/10.1371/journal.pone.0060201
- Primary Citation of Related Structures:
2M01 - PubMed Abstract:
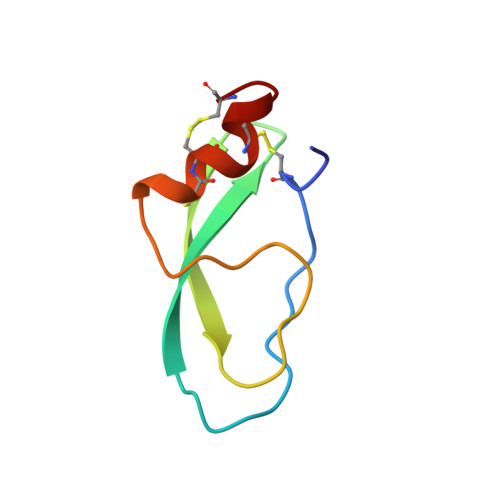
Recently, a new subfamily of long-chain toxins with a Kunitz-type fold was found in scorpion venom glands. Functionally, these toxins inhibit protease activity and block potassium channels. However, the genomic organization and three-dimensional (3-D) structure of this kind of scorpion toxin has not been reported. Here, we characterized the genomic organization and 3-D nuclear magnetic resonance structure of the scorpion Kunitz-type toxin, LmKTT-1a, which has a unique cysteine pattern. The LmKTT-1a gene contained three exons, which were interrupted by two introns located in the mature peptide region. Despite little similarity to other Kunitz-type toxins and a unique pattern of disulfide bridges, LmKTT-1a possessed a conserved Kunitz-type structural fold with one α-helix and two β-sheets. Comparison of the genomic organization, 3-D structure, and functional data of known toxins from the α-KTx, β-KTx, γ-KTx, and κ-KTx subfamily suggested that scorpion Kunitz-type potassium channel toxins might have evolved from a new ancestor that is completely different from the common ancestor of scorpion toxins with a CSα/β fold. Thus, these analyses provide evidence of a new scorpion potassium channel toxin subfamily, which we have named δ-KTx. Our results highlight the genomic, structural, and evolutionary diversity of scorpion potassium channel toxins. These findings may accelerate the design and development of diagnostic and therapeutic peptide agents for human potassium channelopathies.
- State Key Laboratory of Virology, College of Life Sciences, Wuhan University, Wuhan, PR China.
Organizational Affiliation: