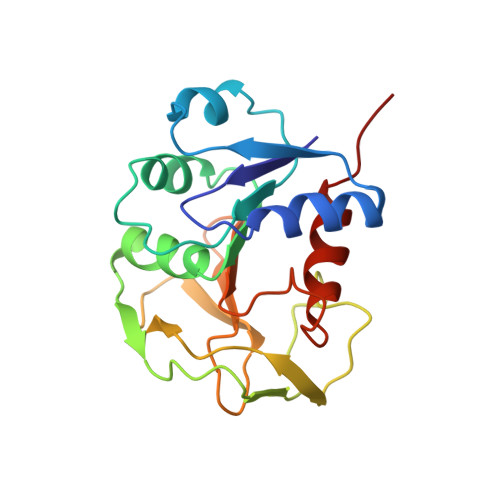
1H, 13C, 15N assignment and secondary structure determination of glutamine amido transferase subunit of gaunosine monophosphate synthetase from Methanocaldococcus jannaschii
Ali, R., Kumar, S., Balaram, H., Sarma, S.P.(2012) Biomol NMR Assign 6: 193-196
- PubMed: 22203461
- DOI: https://doi.org/10.1007/s12104-011-9354-x
- Primary Citation of Related Structures:
2LXN - PubMed Abstract:
Sequence specific resonance assignments have been obtained for (1)H, (13)C and (15)N nuclei of the 21 kDa (188 residues long) glutamine amido transferase subunit of guanosine monophosphate synthetase from Methanocaldococcus jannaschii. From an analysis of (1)H and (13)C(α), (13)C(β) secondary chemical shifts, (3) JH(N)H(α) scalar coupling constants and sequential, short and medium range (1)H-(1)H NOEs, it was deduced that the glutamine amido transferase subunit has eleven strands and five helices as the major secondary structural elements in its tertiary structure.
- 207, Molecular Biophysics Unit, Indian Institute of Science, Bangalore 560012, Karnataka, India.
Organizational Affiliation: