New Tricks of an Old Pattern: STRUCTURAL VERSATILITY OF SCORPION TOXINS WITH COMMON CYSTEINE SPACING.
Saucedo, A.L., Flores-Solis, D., Rodriguez de la Vega, R.C., Ramirez-Cordero, B., Hernandez-Lopez, R., Cano-Sanchez, P., Navarro, R.N., Garcia-Valdes, J., Coronas-Valderrama, F., de Roodt, A., Brieba, L.G., Possani, L.D., Del Rio-Portilla, F.(2012) J Biological Chem 287: 12321-12330
- PubMed: 22238341
- DOI: https://doi.org/10.1074/jbc.M111.329607
- Primary Citation of Related Structures:
2LI3, 2LO7 - PubMed Abstract:
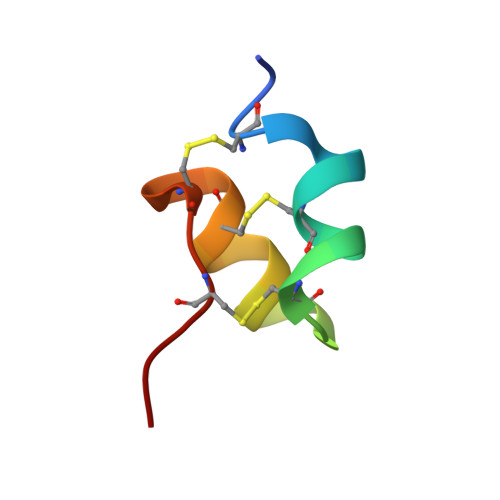
Scorpion venoms are a rich source of K(+) channel-blocking peptides. For the most part, they are structurally related small disulfide-rich proteins containing a conserved pattern of six cysteines that is assumed to dictate their common three-dimensional folding. In the conventional pattern, two disulfide bridges connect an α-helical segment to the C-terminal strand of a double- or triple-stranded β-sheet, conforming a cystine-stabilized α/β scaffold (CSα/β). Here we show that two K(+) channel-blocking peptides from Tityus scorpions conserve the cysteine spacing of common scorpion venom peptides but display an unconventional disulfide pattern, accompanied by a complete rearrangement of the secondary structure topology into a CS helix-loop-helix fold. Sequence and structural comparisons of the peptides adopting this novel fold suggest that it would be a new elaboration of the widespread CSα/β scaffold, thus revealing an unexpected structural versatility of these small disulfide-rich proteins. Acknowledgment of such versatility is important to understand how venom structural complexity emerged on a limited number of molecular scaffolds.
- Instituto de Química, Universidad Nacional Autónoma de México, Ciudad Universitaria, México, D.F., 04510, México.
Organizational Affiliation: