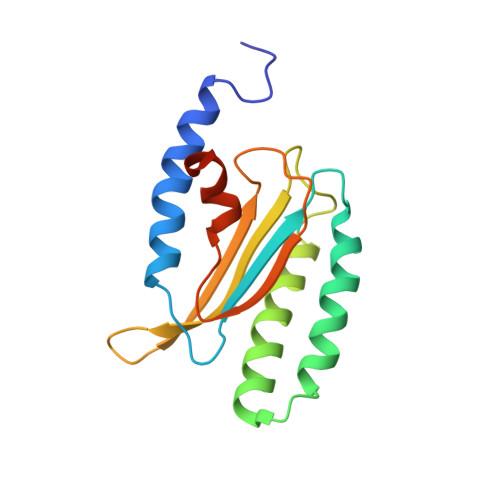
Structure and dynamics in solution of the stop codon decoding N-terminal domain of the human polypeptide chain release factor eRF1.
Polshakov, V.I., Eliseev, B.D., Birdsall, B., Frolova, L.Y.(2012) Protein Sci 21: 896-903
- PubMed: 22517631
- DOI: https://doi.org/10.1002/pro.2067
- Primary Citation of Related Structures:
2LLX - PubMed Abstract:
The high-resolution NMR structure of the N-domain of human eRF1, responsible for stop codon recognition, has been determined in solution. The overall fold of the protein is the same as that found in the crystal structure. However, the structures of several loops, including those participating in stop codon decoding, are different. Analysis of the NMR relaxation data reveals that most of the regions with the highest structural discrepancy between the solution and solid states undergo internal motions on the ps-ns and ms time scales. The NMR data show that the N-domain of human eRF1 exists in two conformational states. The distribution of the residues having the largest chemical shift differences between the two forms indicates that helices α2 and α3, with the NIKS loop between them, can switch their orientation relative to the β-core of the protein. Such structural plasticity may be essential for stop codon recognition by human eRF1.
- Center for Magnetic Tomography and Spectroscopy, Faculty of Fundamental Medicine, MV Lomonosov Moscow State University, Moscow, Russia. vpolsha@mail.ru
Organizational Affiliation: