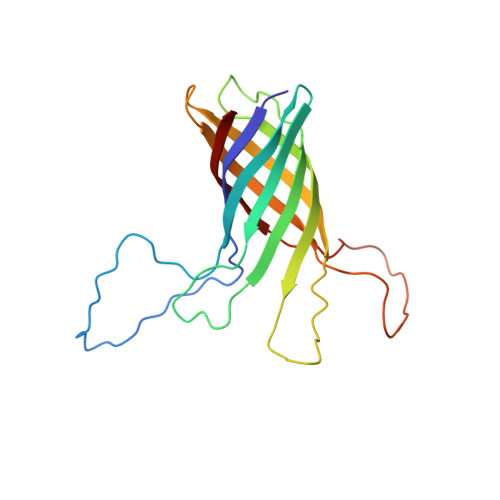
Structural Basis for the Interaction of Lipopolysaccharide with Outer Membrane Protein H (OprH) from Pseudomonas aeruginosa.
Edrington, T.C., Kintz, E., Goldberg, J.B., Tamm, L.K.(2011) J Biological Chem 286: 39211-39223
- PubMed: 21865172
- DOI: https://doi.org/10.1074/jbc.M111.280933
- Primary Citation of Related Structures:
2LHF - PubMed Abstract:
Pseudomonas aeruginosa is a major nosocomial pathogen that infects cystic fibrosis and immunocompromised patients. The impermeability of the P. aeruginosa outer membrane contributes substantially to the notorious antibiotic resistance of this human pathogen. This impermeability is partially imparted by the outer membrane protein H (OprH). Here we have solved the structure of OprH in a lipid environment by solution NMR. The structure reveals an eight-stranded β-barrel protein with four extracellular loops of unequal size. Fast time-scale dynamics measurements show that the extracellular loops are disordered and unstructured. It was previously suggested that the function of OprH is to provide increased stability to the outer membranes of P. aeruginosa by directly interacting with lipopolysaccharide (LPS) molecules. Using in vivo and in vitro biochemical assays, we show that OprH indeed interacts with LPS in P. aeruginosa outer membranes. Based upon NMR chemical shift perturbations observed upon the addition of LPS to OprH in lipid micelles, we conclude that the interaction is predominantly electrostatic and localized to charged regions near both rims of the barrel, but also through two conspicuous tyrosines in the middle of the bilayer. These results provide the first molecular structure of OprH and offer evidence for multiple interactions between OprH and LPS that likely contribute to the antibiotic resistance of P. aeruginosa.
- Center for Membrane Biology, University of Virginia, Charlottesville, Virginia 22908, USA.
Organizational Affiliation: