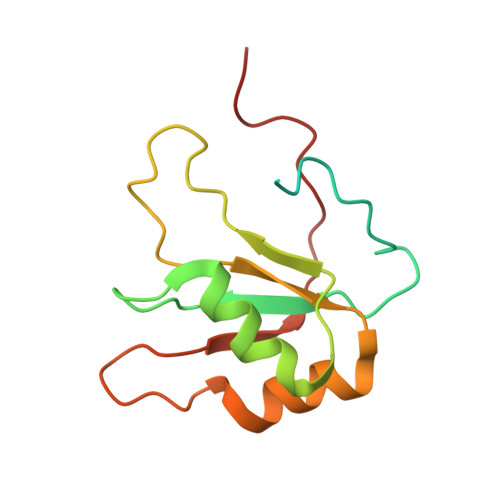
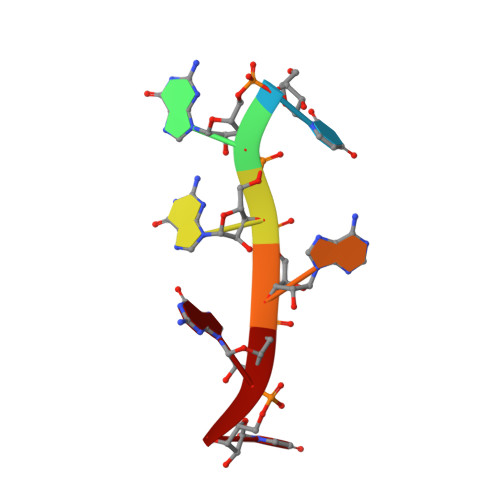
A syn-anti conformational difference allows SRSF2 to recognize guanines and cytosines equally well.
Daubner, G.M., Clery, A., Jayne, S., Stevenin, J., Allain, F.H.(2012) EMBO J 31: 162-174
- PubMed: 22002536
- DOI: https://doi.org/10.1038/emboj.2011.367
- Primary Citation of Related Structures:
2LEA, 2LEB, 2LEC - PubMed Abstract:
SRSF2 (SC35) is a key player in the regulation of alternative splicing events and binds degenerated RNA sequences with similar affinity in nanomolar range. We have determined the solution structure of the SRSF2 RRM bound to the 5'-UCCAGU-3' and 5'-UGGAGU-3' RNA, both identified as SRSF2 binding sites in the HIV-1 tat exon 2. RNA recognition is achieved through a novel sandwich-like structure with both termini forming a positively charged cavity to accommodate the four central nucleotides. To bind both RNA sequences equally well, SRSF2 forms a nearly identical network of intermolecular interactions by simply flipping the bases of the two consecutive C or G nucleotides into either anti or syn conformation. We validate this unusual mode of RNA recognition functionally by in-vitro and in-vivo splicing assays and propose a 5'-SSNG-3' (S=C/G) high-affinity binding consensus sequence for SRSF2. In conclusion, in addition to describe for the first time the RNA recognition mode of SRSF2, we provide the precise consensus sequence to identify new putative binding sites for this splicing factor.
- Institute of Molecular Biology and Biophysics, ETH Zürich, Zürich, Switzerland.
Organizational Affiliation: