Novel dimeric structure of phage 29-encoded protein p56: insights into uracil-DNA glycosylase inhibition.
Asensio, J.L., Perez-Lago, L., Lazaro, J.M., Gonzalez, C., Serrano-Heras, G., Salas, M.(2011) Nucleic Acids Res 39: 9779-9788
- PubMed: 21890898
- DOI: https://doi.org/10.1093/nar/gkr667
- Primary Citation of Related Structures:
2LE2 - PubMed Abstract:
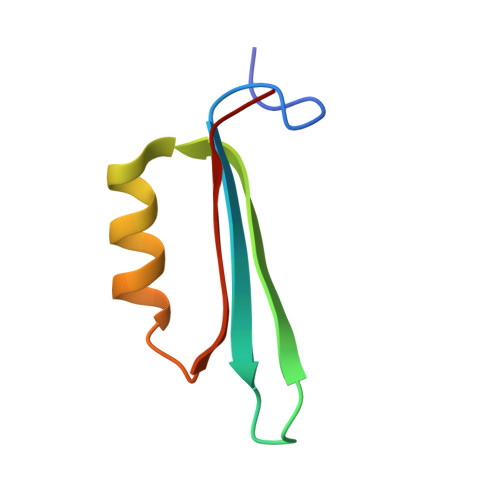
Protein p56 encoded by the Bacillus subtilis phage φ29 inhibits the host uracil-DNA glycosylase (UDG) activity. To get insights into the structural basis for this inhibition, the NMR solution structure of p56 has been determined. The inhibitor defines a novel dimeric fold, stabilized by a combination of polar and extensive hydrophobic interactions. Each polypeptide chain contains three stretches of anti-parallel β-sheets and a helical region linked by three short loops. In addition, microcalorimetry titration experiments showed that it forms a tight 2:1 complex with UDG, strongly suggesting that the dimer represents the functional form of the inhibitor. This was further confirmed by the functional analysis of p56 mutants unable to assemble into dimers. We have also shown that the highly anionic region of the inhibitor plays a significant role in the inhibition of UDG. Thus, based on these findings and taking into account previous results that revealed similarities between the association mode of p56 and the phage PBS-1/PBS-2-encoded inhibitor Ugi with UDG, we propose that protein p56 might inhibit the enzyme by mimicking its DNA substrate.
- Departamento de Química Orgánica Biológica, Instituto de Química Orgánica General, CSIC, 28006 Madrid, Spain. iqoa110@iqog.csic.es
Organizational Affiliation: