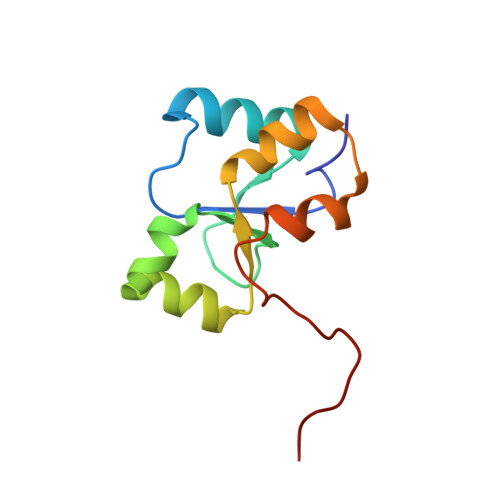
Structural studies of the PARP-1 BRCT domain.
Loeffler, P.A., Cuneo, M.J., Mueller, G.A., Derose, E.F., Gabel, S.A., London, R.E.(2011) BMC Struct Biol 11: 37-37
- PubMed: 21967661
- DOI: https://doi.org/10.1186/1472-6807-11-37
- Primary Citation of Related Structures:
2LE0 - PubMed Abstract:
Poly(ADP-ribose) polymerase-1 (PARP-1) is one of the first proteins localized to foci of DNA damage. Upon activation by encountering nicked DNA, the PARP-1 mediated trans-poly(ADP-ribosyl)ation of DNA binding proteins occurs, facilitating access and accumulation of DNA repair factors. PARP-1 also auto-(ADP-ribosyl)ates its central BRCT-containing domain forming part of an interaction site for the DNA repair scaffolding protein X-ray cross complementing group 1 protein (XRCC1). The co-localization of XRCC1, as well as bound DNA repair factors, to sites of DNA damage is important for cell survival and genomic integrity. Here we present the solution structure and biophysical characterization of the BRCT domain of rat PARP-1. The PARP-1 BRCT domain has the globular α/β fold characteristic of BRCT domains and has a thermal melting transition of 43.0°C. In contrast to a previous characterization of this domain, we demonstrate that it is monomeric in solution using both gel-filtration chromatography and small-angle X-ray scattering. Additionally, we report that the first BRCT domain of XRCC1 does not interact significantly with the PARP-1 BRCT domain in the absence of ADP-ribosylation. Moreover, none of the interactions with other longer PARP-1 constructs which previously had been demonstrated in a pull-down assay of mammalian cell extracts were detected. The PARP-1 BRCT domain has the conserved BRCT fold that is known to be an important protein:protein interaction module in DNA repair and cell signalling pathways. Data indicating no significant protein:protein interactions between PARP-1 and XRCC1 likely results from the absence of poly(ADP-ribose) in one or both binding partners, and further implicates a poly(ADP-ribose)-dependent mechanism for localization of XRCC1 to sites of DNA damage.
- Department of Chemistry, Sam Houston State University, Huntsville, Texas 77340, USA.
Organizational Affiliation: