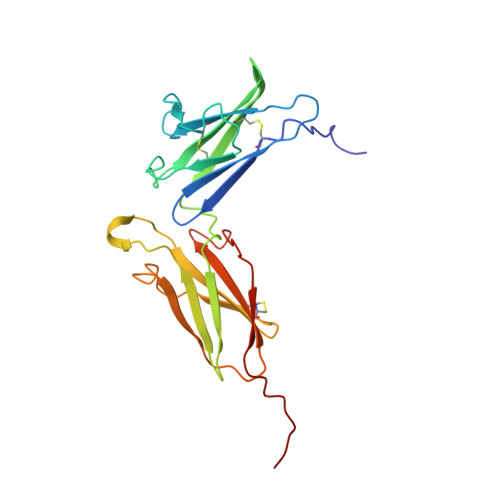
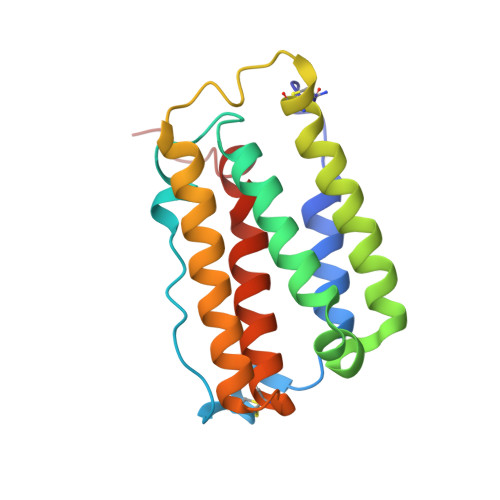
Observation of Intermolecular Interactions in Large Protein Complexes by 2D-Double Difference Nuclear Overhauser Enhancement Spectroscopy: Application to the 44 kDa Interferon-Receptor Complex.
Nudelman, I., Akabayov, S.R., Scherf, T., Anglister, J.(2011) J Am Chem Soc 133: 14755-14764
- PubMed: 21819146
- DOI: https://doi.org/10.1021/ja205480v
- Primary Citation of Related Structures:
2LAG - PubMed Abstract:
NMR detection of intermolecular interactions between protons in large protein complexes is very challenging because it is difficult to distinguish between weak NOEs from intermolecular interactions and the much larger number of strong intramolecular NOEs. This challenging task is exacerbated by the decrease in signal-to-noise ratio in the often used isotope-edited and isotope-filtered experiments as a result of enhanced T(2) relaxation. Here, we calculate a double difference spectrum that shows exclusively intermolecular NOEs and manifests the good signal-to-noise ratio in 2D homonuclear NOESY spectra even for large proteins. The method is straightforward and results in a complete picture of all intermolecular interactions involving non exchangeable protons. Ninety-seven such (1)H-(1)H NOEs were assigned for the 44 KDa interferon-α2/IFNAR2 complex and used for docking these two proteins. The symmetry of the difference spectrum, its superb resolution, and unprecedented signal-to-noise ratio in this large protein/receptor complex suggest that this method is generally applicable to study large biopolymeric complexes.
- Department of Structural Biology, Weizmann Institute of Science, Rehovot 76100, Israel.
Organizational Affiliation: