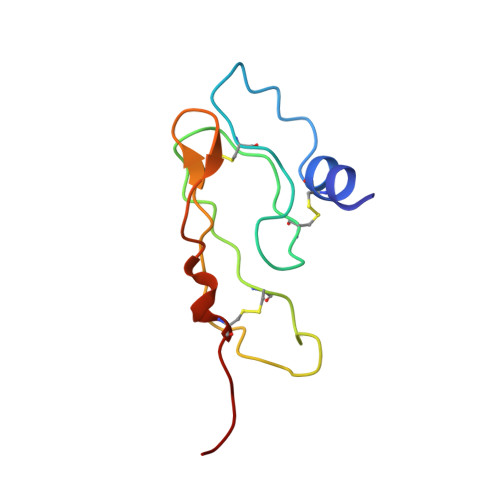
NMR structure of the first extracellular domain of corticotropin-releasing factor receptor 1 (ECD1-CRF-R1) complexed with a high affinity agonist.
Grace, C.R., Perrin, M.H., Gulyas, J., Rivier, J.E., Vale, W.W., Riek, R.(2010) J Biological Chem 285: 38580-38589
- PubMed: 20843795
- DOI: https://doi.org/10.1074/jbc.M110.121897
- Primary Citation of Related Structures:
2L27 - PubMed Abstract:
The corticotropin-releasing factor (CRF) peptide hormone family members coordinate endocrine, behavioral, autonomic, and metabolic responses to stress and play important roles within the cardiovascular, gastrointestinal, and central nervous systems, among others. The actions of the peptides are mediated by activation of two G-protein-coupled receptors of the B1 family, CRF receptors 1 and 2 (CRF-R1 and CRF-R2α,β). The recently reported three-dimensional structures of the first extracellular domain (ECD1) of both CRF-R1 and CRF-R2β (Pioszak, A. A., Parker, N. R., Suino-Powell, K., and Xu, H. E. (2008) J. Biol. Chem. 283, 32900-32912; Grace, C. R., Perrin, M. H., Gulyas, J., Digruccio, M. R., Cantle, J. P., Rivier, J. E., Vale, W. W., and Riek, R. (2007) Proc. Natl. Acad. Sci. U.S.A. 104, 4858-4863) complexed with peptide antagonists provided a starting point in understanding the binding between CRF ligands and receptors at a molecular level. We now report the three-dimensional NMR structure of the ECD1 of human CRF-R1 complexed with a high affinity agonist, α-helical cyclic CRF. In the structure of the complex, the C-terminal residues (23-41) of α-helical cyclic CRF bind to the ECD1 of CRF-R1 in a helical conformation mainly along the hydrophobic face of the peptide in a manner similar to that of the antagonists in their corresponding ECD1 complex structures. Unique to this study is the observation that complex formation between an agonist and the ECD1-CRF-R1 promotes the helical conformation of the N terminus of the former, important for receptor activation (Gulyas, J., Rivier, C., Perrin, M., Koerber, S. C., Sutton, S., Corrigan, A., Lahrichi, S. L., Craig, A. G., Vale, W., and Rivier, J. (1995) Proc. Natl. Acad. Sci. U.S.A. 92, 10575-10579).
- Structural Biology Laboratory, Salk Institute for Biological Studies, La Jolla, California 92037, USA.
Organizational Affiliation: