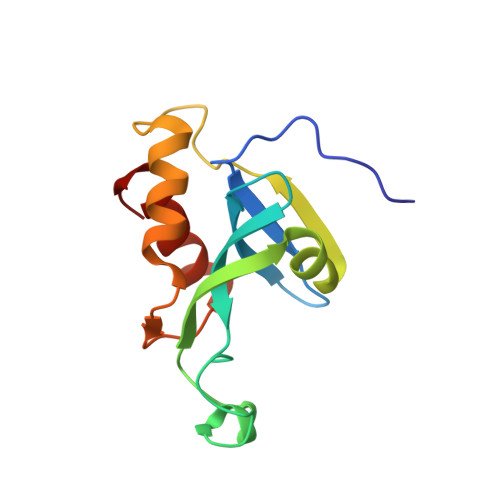
Solution structure of the N-terminal catalytic domain of human H-REV107--a novel circular permutated NlpC/P60 domain
Ren, X., Lin, J., Jin, C., Xia, B.(2010) FEBS Lett 584: 4222-4226
- PubMed: 20837014
- DOI: https://doi.org/10.1016/j.febslet.2010.09.015
- Primary Citation of Related Structures:
2KYT - PubMed Abstract:
H-REV107 is a Ca(2+)-independent phospholipase A(1/2), and it is also a pro-apoptosis protein belonging to the novel class II tumor suppressor family, H-REV107-like family. Here we report the solution structure of the N-terminal catalytic domain of human H-REV107, which has a similar architecture to classical NlpC/P60 domains, even though their fold topologies are different due to circular permutation in the primary sequence. The phospholipase active site possesses a structurally conserved Cys-His-His catalytic triad as found in NlpC/P60 peptidases, indicating H-REV107 should adopt a similar catalytic mechanism towards phospholipid substrates to that of NlpC/P60 peptidases towards peptides. As H-REV107 is highly similar to lecithin retinol acyltransferase, our study also provides structural insight to this essential enzyme in retinol metabolism.
- Beijing Nuclear Magnetic Resonance Center, Peking University, Beijing, China.
Organizational Affiliation: