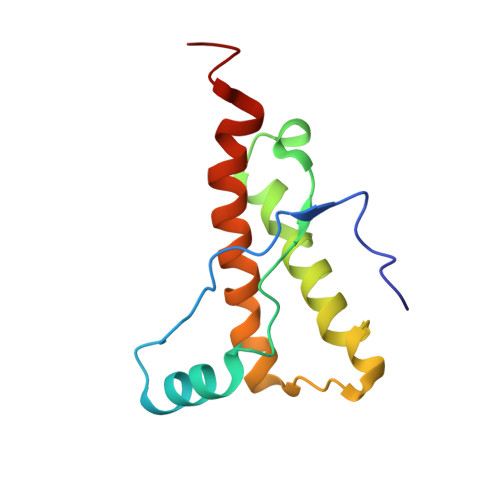
Horse prion protein NMR structure and comparisons with related variants of the mouse prion protein.
Perez, D.R., Damberger, F.F., Wuthrich, K.(2010) J Mol Biology 400: 121-128
- PubMed: 20460128
- DOI: https://doi.org/10.1016/j.jmb.2010.04.066
- Primary Citation of Related Structures:
2KU4, 2KU5, 2KU6 - PubMed Abstract:
The NMR structure of the horse (Equus caballus) cellular prion protein at 25 degrees C exhibits the typical PrP(C) [cellular form of prion protein (PrP)] global architecture, but in contrast to most other mammalian PrP(C)s, it contains a well-structured loop connecting the beta2 strand with the alpha2 helix. Comparison with designed variants of the mouse prion protein resulted in the identification of a single amino acid exchange within the loop, D167S, which correlates with the high structural order of this loop in the solution structure at 25 degrees C and is unique to the PrP sequences of equine species. The beta2-alpha2 loop and the alpha3 helix form a protein surface epitope that has been proposed to be the recognition area for a hypothetical chaperone, "protein X," which would promote conversion of PrP(C) into the disease-related scrapie form and thus mediate intermolecular interactions related to the transmission barrier for transmissible spongiform encephalopathies (TSEs) between different species. The present results are evaluated in light of recent indications from in vivo experiments that the local beta2-alpha2 loop structure affects the susceptibility of transgenic mice to TSEs and the fact that there are no reports on TSE in horses.
- Institute of Molecular Biology and Biophysics, ETH Zurich, Schafmattstrasse 20, CH-8093 Zurich, Switzerland.
Organizational Affiliation: