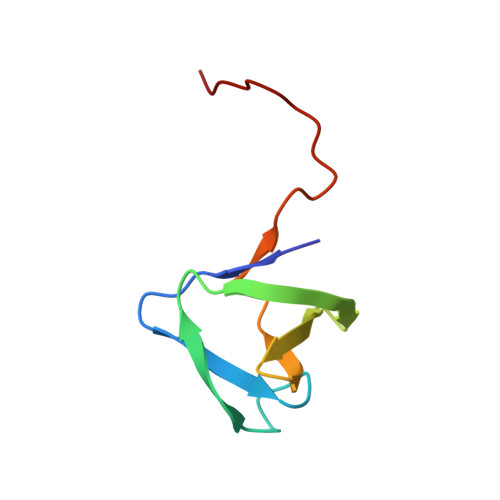
Solution NMR structure of SH3 domain from CPF_0587 (fragment 415-479) from Clostridium perfringens. Northeast Structural Genomics Consortium (NESG) Target CpR74A.
Ramelot, T.A., Cort, J.R., Maglaqui, M., Ciccosanti, C., Janjua, H., Nair, R., Rost, B., Acton, T.B., Xiao, R., Everett, J.K., Montelione, G.T., Kennedy, M.A.To be published.