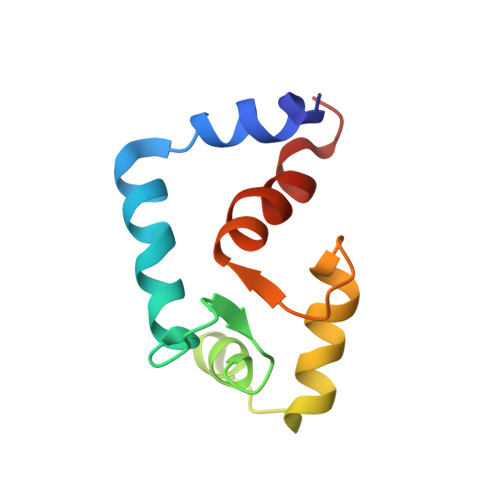
Solution structure of the regulatory domain of human cardiac troponin C in complex with the switch region of cardiac troponin I and W7: the basis of W7 as an inhibitor of cardiac muscle contraction.
Oleszczuk, M., Robertson, I.M., Li, M.X., Sykes, B.D.(2010) J Mol Cell Cardiol 48: 925-933
- PubMed: 20116385
- DOI: https://doi.org/10.1016/j.yjmcc.2010.01.016
- Primary Citation of Related Structures:
2KRD - PubMed Abstract:
The solution structure of Ca(2+)-bound regulatory domain of cardiac troponin C (cNTnC) in complex with the switch region of troponin I (cTnI(147-163)) and the calmodulin antagonist, N-(6-aminohexyl)-5-chloro-1-naphthalenesulfinamide (W7), has been determined by NMR spectroscopy. The structure reveals that the W7 naphthalene ring interacts with the terminal methyl groups of M47, M60, and M81 as well as aliphatic and aromatic side chains of several other residues in the hydrophobic pocket of cNTnC. The H3 ring proton of W7 also contacts the methyl groups of I148 and M153 of cTnI(147-163). The N-(6-aminohexyl) tail interacts primarily with the methyl groups of V64 and M81, which are located on the C- and D-helices of cNTnC. Compared to the structure of the cNTnC*Ca(2+)*W7 complex (Hoffman, R. M. B. and Sykes, B. D. (2009) Biochemistry 48, 5541-5552), the tail of W7 reorients slightly toward the surface of cNTnC while the ring remains in the hydrophobic pocket. The positively charged -NH(3)(+) group from the tail of W7 repels the positively charged R147 of cTnI(147-163). As a result, the N-terminus of the peptide moves away from cNTnC and the helical content of cTnI(147-163) is diminished, when compared to the structure of cNTnC*Ca(2+)*cTnI(147-163) (Li, M. X., Spyracopoulos, L., and Sykes B. D. (1999) Biochemistry 38, 8289-8298). Thus the ternary structure cNTnC*Ca(2+)*W7*cTnI(147-163) reported in this study offers an explanation for the approximately 13-fold affinity reduction of cTnI(147-163) for cNTnC*Ca(2+) in the presence of W7 and provides a structural basis for the inhibitory effect of W7 in cardiac muscle contraction. This generates molecular insight into structural features that are useful for the design of cTnC-specific Ca(2+)-desensitizing drugs.
- Department of Biochemistry, School of Molecular and Systems Medicine, University of Alberta, Edmonton, Alberta, Canada T6G 2H7.
Organizational Affiliation: