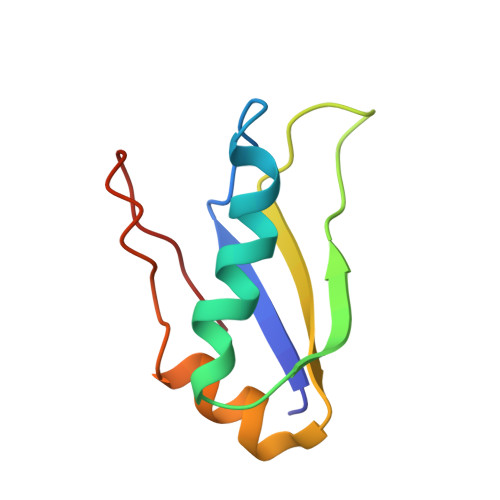
The indispensable N-terminal half of eIF3j/HCR1 co-operates with its structurally conserved binding partner eIF3b/PRT1-RRM and eIF1A in stringent AUG selection
Elantak, L., Wagner, S., Herrmannova, A., Janoskova, M., Rutkai, E., Lukavsky, P.J., Valasek, L.To be published.