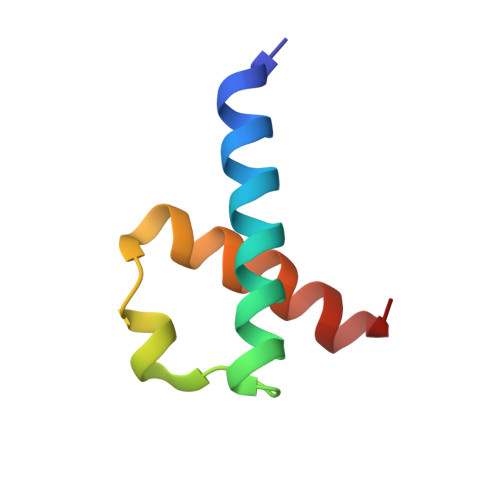
The N-terminus of the human RecQL4 helicase is a homeodomain-like DNA interaction motif
Ohlenschlager, O., Kuhnert, A., Schneider, A., Haumann, S., Bellstedt, P., Keller, H., Saluz, H.P., Hortschansky, P., Hanel, F., Grosse, F., Gorlach, M., Pospiech, H.(2012) Nucleic Acids Res 40: 8309-8324
- PubMed: 22730300
- DOI: https://doi.org/10.1093/nar/gks591
- Primary Citation of Related Structures:
2KMU - PubMed Abstract:
The RecQL4 helicase is involved in the maintenance of genome integrity and DNA replication. Mutations in the human RecQL4 gene cause the Rothmund-Thomson, RAPADILINO and Baller-Gerold syndromes. Mouse models and experiments in human and Xenopus have proven the N-terminal part of RecQL4 to be vital for cell growth. We have identified the first 54 amino acids of RecQL4 (RecQL4_N54) as the minimum interaction region with human TopBP1. The solution structure of RecQL4_N54 was determined by heteronuclear liquid-state nuclear magnetic resonance (NMR) spectroscopy (PDB 2KMU; backbone root-mean-square deviation 0.73 Å). Despite low-sequence homology, the well-defined structure carries an overall helical fold similar to homeodomain DNA-binding proteins but lacks their archetypical, minor groove-binding N-terminal extension. Sequence comparison indicates that this N-terminal homeodomain-like fold is a common hallmark of metazoan RecQL4 and yeast Sld2 DNA replication initiation factors. RecQL4_N54 binds DNA without noticeable sequence specificity yet with apparent preference for branched over double-stranded (ds) or single-stranded (ss) DNA. NMR chemical shift perturbation observed upon titration with Y-shaped, ssDNA and dsDNA shows a major contribution of helix α3 to DNA binding, and additional arginine side chain interactions for the ss and Y-shaped DNA.
- Research Group Biomolecular NMR Spectroscopy, Leibniz Institute for Age Research-Fritz Lipmann Institute, Beutenbergstr. 11, Jena, Germany.
Organizational Affiliation: