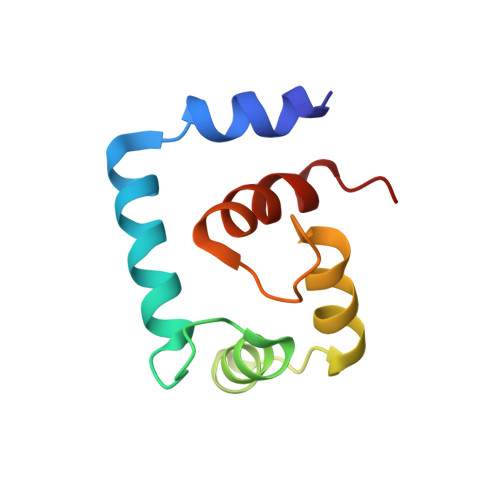
The effect of the cosolvent trifluoroethanol on a tryptophan side chain orientation in the hydrophobic core of troponin C.
Julien, O., Mercier, P., Crane, M.L., Sykes, B.D.(2009) Protein Sci 18: 1165-1174
- PubMed: 19472326
- DOI: https://doi.org/10.1002/pro.121
- Primary Citation of Related Structures:
2KGB - PubMed Abstract:
The unique biophysical properties of tryptophan residues have been exploited for decades to monitor protein structure and dynamics using a variety of spectroscopic techniques, such as fluorescence and nuclear magnetic resonance (NMR). We recently designed a tryptophan mutant in the regulatory N-domain of cardiac troponin C (F77W-cNTnC) to study the domain orientation of troponin C in muscle fibers using solid-state NMR. In our previous study, we determined the NMR structure of calcium-saturated mutant F77W-V82A-cNTnC in the presence of 19% 2,2,2-trifluoroethanol (TFE). TFE is a widely used cosolvent in the biophysical characterization of the solution structures of peptides and proteins. It is generally assumed that the structures are unchanged in the presence of cosolvents at relatively low concentrations, and this has been verified for TFE at the level of the overall secondary and tertiary structure for several calcium regulatory proteins. Here, we present the NMR solution structure of the calcium saturated F77W-cNTnC in presence of its biological binding partner troponin I peptide (cTnI(144-163)) and in the absence of TFE. We have also characterized a panel of six F77W-cNTnC structures in the presence and absence TFE, cTnI(144-163), and the extra mutation V82A, and used (19)F NMR to characterize the effect of TFE on the F77(5fW) analog. Our results show that although TFE did not perturb the overall protein structure, TFE did induce a change in the orientation of the indole ring of the buried tryptophan side chain from the anticipated position based upon homology with other proteins, highlighting the potential dangers of the use of cosolvents.
- Department of Biochemistry, University of Alberta, Edmonton, Alberta, Canada.
Organizational Affiliation: