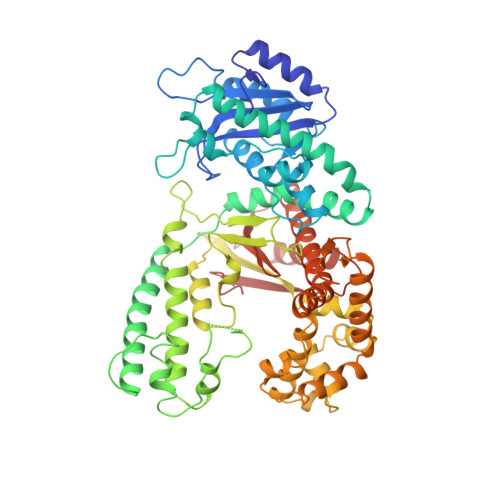
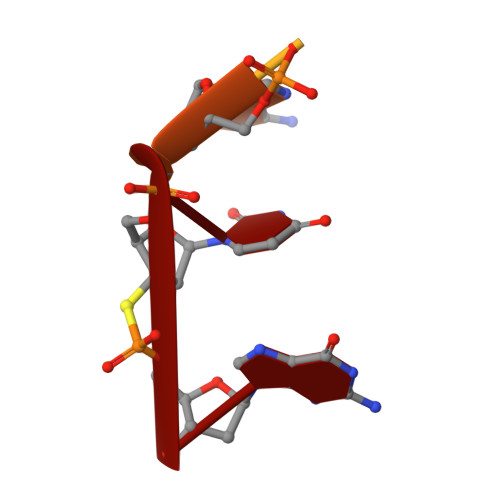
Structures of normal single-stranded DNA and deoxyribo-3'-S-phosphorothiolates bound to the 3'-5' exonucleolytic active site of DNA polymerase I from Escherichia coli.
Brautigam, C.A., Sun, S., Piccirilli, J.A., Steitz, T.A.(1999) Biochemistry 38: 696-704
- PubMed: 9888810
- DOI: https://doi.org/10.1021/bi981537g
- Primary Citation of Related Structures:
2KFN, 2KFZ, 2KZM, 2KZZ - PubMed Abstract:
The interaction of a divalent metal ion with a leaving 3' oxygen is a central component of several proposed mechanisms of phosphoryl transfer. In support of this are recent kinetic studies showing that thiophilic metal ions (e.g., Mn2+) stimulate the hydrolysis of compounds in which sulfur takes the place of the leaving oxygen. To examine the structural basis of this phenomenon, we have solved four crystal structures of single-stranded DNA's containing either oxygen or sulfur at a 3'-bridging position bound in conjunction with various metal ions at the 3'-5' exonucleolytic active site of the Klenow fragment (KF) of DNA polymerase I from Escherichia coli. Two structures of normal ssDNA bound to KF in the presence of Zn2+ and Mn2+ or Zn2+ alone were refined at 2.6- and 2.25-A resolution, respectively. They serve as standards for comparison with other Mn2+- and Zn2+-containing structures. In these cases, Mn2+ and Zn2+ bind at metal ion site B in a nearly identical position to Mg2+ (Brautigam and Steitz (1998) J. Mol. Biol. 277, 363-377). Two structures of KF bound to a deoxyoligonucleotide that contained a 3'-bridging sulfur at the scissile phosphate were refined at 2.03-A resolution. Although the bridging sulfur compounds bind in a manner very similar to that of the normal oligonucleotides, the presence of the sulfur changes the metal ion binding properties of the active site such that Mn2+ and Zn2+ are observed at metal ion site B, but Mg2+ is not. It therefore appears that the ability of the bridging sulfur compounds to exclude nonthiophilic metal ions from metal ion site B explains the low activity of KF exonuclease on these substrates in the presence of Mg2+ (Curley et al. (1997) J. Am. Chem. Soc. 119, 12691-12692) and that the 3'-bridging atom of the substrate is influencing the binding of metal ion B prior to catalysis.
- Department of Molecular Biophysics & Biochemistry, Howard Hughes Medical Institute, Yale University, New Haven, Connecticut 06520-8114, USA.
Organizational Affiliation: