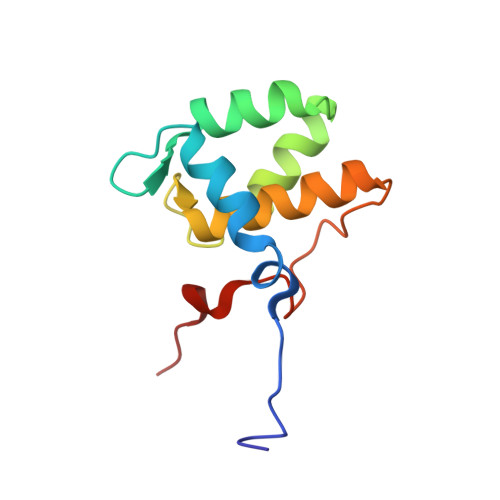
Structural insight into the interaction of proteins containing NPF, DPF, and GPF motifs with the C-terminal EH-domain of EHD1.
Kieken, F., Jovic, M., Tonelli, M., Naslavsky, N., Caplan, S., Sorgen, P.L.(2009) Protein Sci 18: 2471-2479
- PubMed: 19798736
- DOI: https://doi.org/10.1002/pro.258
- Primary Citation of Related Structures:
2KFF, 2KFG, 2KFH - PubMed Abstract:
Eps15 homology (EH)-domain containing proteins are regulators of endocytic membrane trafficking. EH-domain binding to proteins containing the tripeptide NPF has been well characterized, but recent studies have shown that EH-domains are also able to interact with ligands containing DPF or GPF motifs. We demonstrate that the three motifs interact in a similar way with the EH-domain of EHD1, with the NPF motif having the highest affinity due to the presence of an intermolecular hydrogen bond. The weaker affinity for the DPF and GPF motifs suggests that if complex formation occurs in vivo, they may require high ligand concentrations, the presence of successive motifs and/or specific flanking residues.
- Department of Biochemistry and Molecular Biology and Eppley Cancer Center, University of Nebraska Medical Center, Omaha, Nebraska 68198-5870, USA.
Organizational Affiliation: