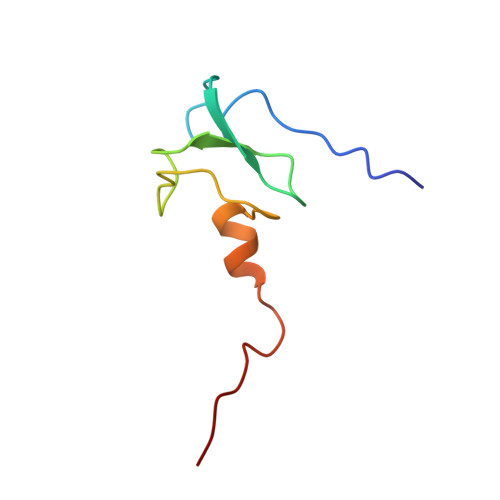
The solution structure of the first PHD finger of autoimmune regulator in complex with non-modified histone H3 tail reveals the antagonistic role of H3R2 methylation
Chignola, F., Gaetani, M., Rebane, A., Org, T., Mollica, L., Zucchelli, C., Spitaleri, A., Mannella, V., Peterson, P., Musco, G.(2009) Nucleic Acids Res 37: 2951-2961
- PubMed: 19293276
- DOI: https://doi.org/10.1093/nar/gkp166
- Primary Citation of Related Structures:
2KE1 - PubMed Abstract:
Plant homeodomain (PHD) fingers are often present in chromatin-binding proteins and have been shown to bind histone H3 N-terminal tails. Mutations in the autoimmune regulator (AIRE) protein, which harbours two PHD fingers, cause a rare monogenic disease, autoimmune polyendocrinopathy-candidiasis-ectodermal dystrophy (APECED). AIRE activates the expression of tissue-specific antigens by directly binding through its first PHD finger (AIRE-PHD1) to histone H3 tails non-methylated at K4 (H3K4me0). Here, we present the solution structure of AIRE-PHD1 in complex with H3K4me0 peptide and show that AIRE-PHD1 is a highly specialized non-modified histone H3 tail reader, as post-translational modifications of the first 10 histone H3 residues reduce binding affinity. In particular, H3R2 dimethylation abrogates AIRE-PHD1 binding in vitro and reduces the in vivo activation of AIRE target genes in HEK293 cells. The observed antagonism by R2 methylation on AIRE-PHD1 binding is unique among the H3K4me0 histone readers and represents the first case of epigenetic negative cross-talk between non-methylated H3K4 and methylated H3R2. Collectively, our results point to a very specific histone code responsible for non-modified H3 tail recognition by AIRE-PHD1 and describe at atomic level one crucial step in the molecular mechanism responsible for antigen expression in the thymus.
- Biomolecular NMR Laboratory, Dulbecco Telethon Institute, Milan, Italy.
Organizational Affiliation: