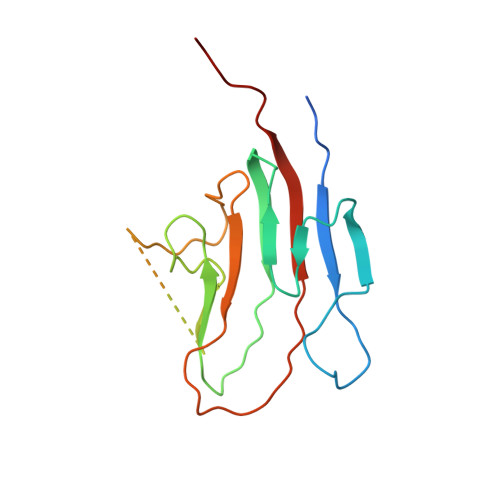
NMR-based homology model for the solution structure of the C-terminal globular domain of EMILIN1
Verdone, G., Corazza, A., Colebrooke, S.A., Cicero, D., Eliseo, T., Boyd, J., Doliana, R., Fogolari, F., Viglino, P., Colombatti, A., Campbell, I.D., Esposito, G.(2009) J Biomol NMR 43: 79-96
- PubMed: 19023665
- DOI: https://doi.org/10.1007/s10858-008-9290-y
- Primary Citation of Related Structures:
2KA3 - PubMed Abstract:
EMILIN1 is a glycoprotein of elastic tissues that has been recently linked to the pathogenesis of hypertension. The protein is formed by different independently folded structural domains whose role has been partially elucidated. In this paper the solution structure, inferred from NMR-based homology modelling of the C-terminal trimeric globular C1q domain (gC1q) of EMILIN1, is reported. The high molecular weight and the homotrimeric structure of the protein required the combined use of highly deuterated (15)N, (13)C-labelled samples and TROSY experiments. Starting from a homology model, the protein structure was refined using heteronuclear residual dipolar couplings, chemical shift patterns, NOEs and H-exchange data. Analysis of the gC1q domain structure of EMILIN1 shows that each protomer of the trimer adopts a nine-stranded beta sandwich folding topology which is related to the conformation observed for other proteins of the family. Distinguishing features, however, include a missing edge-strand and an unstructured 19-residue loop. Although the current data do not allow this loop to be precisely defined, the available evidence is consistent with a flexible segment that protrudes from each subunit of the globular trimeric assembly and plays a key role in inter-molecular interactions between the EMILIN1 gC1q homotrimer and its integrin receptor alpha4beta1.
- Dipartimento di Scienze e Tecnologie Biomediche-MATI Centre of Excellence, Università di Udine, P. le Kolbe, 4-33100, Udine, Italy.
Organizational Affiliation: