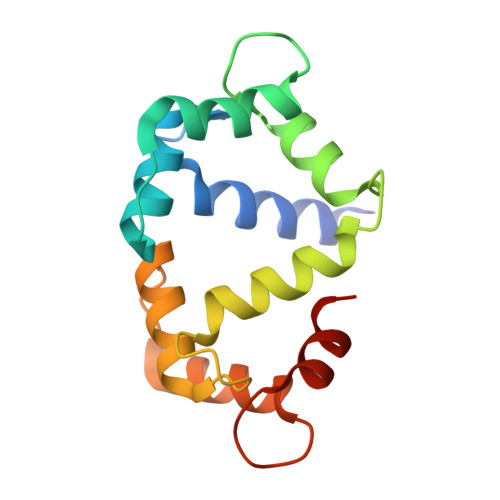
A coupled equilibrium shift mechanism in calmodulin-mediated signal transduction
Gsponer, J., Christodoulou, J., Cavalli, A., Bui, J.M., Richter, B., Dobson, C.M., Vendruscolo, M.(2008) Structure 16: 736-746
- PubMed: 18462678
- DOI: https://doi.org/10.1016/j.str.2008.02.017
- Primary Citation of Related Structures:
2K0E, 2K0F - PubMed Abstract:
We used nuclear magnetic resonance data to determine ensembles of conformations representing the structure and dynamics of calmodulin (CaM) in the calcium-bound state (Ca(2+)-CaM) and in the state bound to myosin light chain kinase (CaM-MLCK). These ensembles reveal that the Ca(2+)-CaM state includes a range of structures similar to those present when CaM is bound to MLCK. Detailed analysis of the ensembles demonstrates that correlated motions within the Ca(2+)-CaM state direct the structural fluctuations toward complex-like substates. This phenomenon enables initial ligation of MLCK at the C-terminal domain of CaM and induces a population shift among the substates accessible to the N-terminal domain, thus giving rise to the cooperativity associated with binding. Based on these results and the combination of modern free energy landscape theory with classical allostery models, we suggest that a coupled equilibrium shift mechanism controls the efficient binding of CaM to a wide range of ligands.
- Department of Chemistry, University of Cambridge, Lensfield Road, Cambridge CB2 1EW, United Kingdom.
Organizational Affiliation: