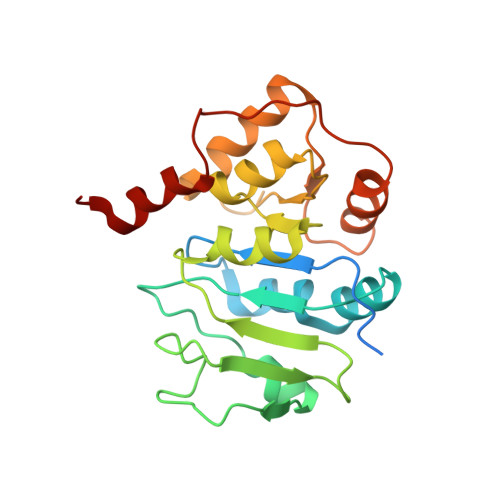
Solution structure of Alg13: the sugar donor subunit of a yeast N-acetylglucosamine transferase.
Wang, X., Weldeghiorghis, T., Zhang, G., Imperiali, B., Prestegard, J.H.(2008) Structure 16: 965-975
- PubMed: 18547528
- DOI: https://doi.org/10.1016/j.str.2008.03.010
- Primary Citation of Related Structures:
2JZC - PubMed Abstract:
The solution structure of Alg13, the glycosyl donor-binding domain of an important bipartite glycosyltransferase in the yeast Saccharomyces cerevisiae, is presented. This glycosyltransferase is unusual in that it is active only in the presence of a binding partner, Alg14. Alg13 is found to adopt a unique topology among glycosyltransferases. Rather than the conventional Rossmann fold found in all GT-B enzymes, the N-terminal half of the protein is a Rossmann-like fold with a mixed parallel and antiparallel beta sheet. The Rossmann fold of the C-terminal half of Alg13 is conserved. However, although conventional GT-B enzymes usually possess three helices at the C terminus, only two helices are present in Alg13. Titration of Alg13 with both UDP-GlcNAc, the native glycosyl donor, and a paramagnetic mimic, UDP-TEMPO, shows that the interaction of Alg13 with the sugar donor is primarily through the residues in the C-terminal half of the protein.
- Complex Carbohydrate Research Center, University of Georgia, Athens, GA 30602, USA.
Organizational Affiliation: