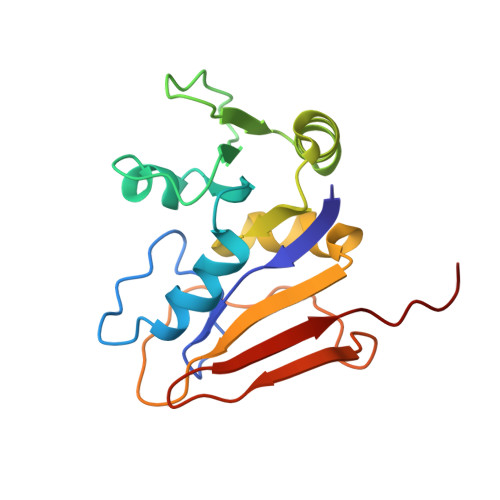
NMR-derived folate-bound structure of dihydrofolate reductase 1 from the halophile Haloferax volcanii.
Boroujerdi, A.F., Young, J.K.(2009) Biopolymers 91: 140-144
- PubMed: 18825778
- DOI: https://doi.org/10.1002/bip.21096
- Primary Citation of Related Structures:
2JYB - PubMed Abstract:
Folate binds to dihydrofolate reductase (DHFR) to form a binary complex whose structure maintains the overall configuration of the enzyme; however, some significant changes are evident when a comparison is made to the enzyme. The structure of DHFR1 from the halophilic Halopherax volcanii was solved in its folate-bound form using nuclear magnetic resonance spectroscopy. NOE data obtained from the (15)N-NOESY-HSQC and (13)C-NOESY-HSQC experiments of the triply labeled ((1)H, (13)C, and (15)N) binary complex were used as input for the structure calculation with the Crystallography and Nuclear Magnetic Resonance System program. The resulting family of structures was compared with the enzyme solved by both nuclear magnetic resonance and X-ray crystallography and also to the mesophilic folate-bound enzyme from Escherichia coli. The binary complex exhibited less convergence of structure in the alpha2-helix and differences in the hinge residues D39 and A94. In comparison to the previously reported mesophilic binary complex solved by X-ray crystallography, the halophilic binary complex reported here does not agree with the convergence of the M20 loop to a single structure. The corresponding L21 loop of the halophilic binary complex family of structures solved by nuclear magnetic resonance indicates variability in this region.
- Department of Chemistry, Mississippi State University, MS 39762, USA.
Organizational Affiliation: