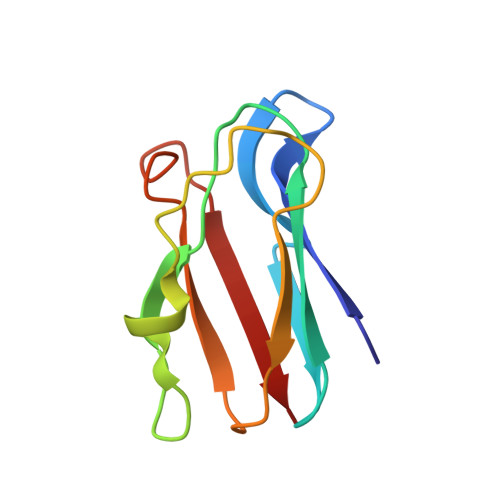
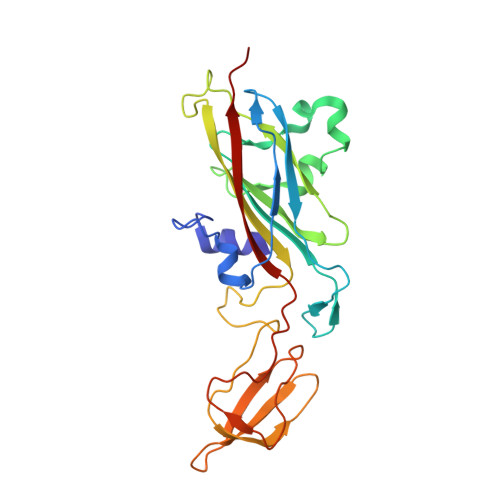
Dynamics in the transient complex of plastocyanin-cytochrome f from Prochlorothrix hollandica.
Hulsker, R., Baranova, M.V., Bullerjahn, G.S., Ubbink, M.(2008) J Am Chem Soc 130: 1985-1991
- PubMed: 18201089
- DOI: https://doi.org/10.1021/ja077453p
- Primary Citation of Related Structures:
2JXM - PubMed Abstract:
The nature of transient protein complexes can range from a highly dynamic ensemble of orientations to a single well-defined state. This represents variation in the equilibrium between the encounter and final, functional state. The transient complex between plastocyanin (Pc) and cytochrome f (cytf) of the cyanobacterium Prochlorothrix hollandica was characterized by NMR spectroscopy. Intermolecular pseudocontact shifts and chemical shift perturbations were used as restraints in docking calculations to determine the structure of the wild-type Pc-cytf complex. The orientation of Pc is similar to orientations found in Pc-cytf complexes from other sources. Electrostatics seems to play a modest role in complex formation. A large variability in the ensemble of lowest energy structures indicates a dynamic nature of the complex. Two unusual hydrophobic patch residues in Pc have been mutated to the residues found in other plastocyanins (Y12G/P14L). The binding constants are similar for the complexes of cytf with wild-type Pc and mutant Pc, but the chemical shift perturbations are smaller for the complex with mutant Pc. Docking calculations for the Y12G/P14L Pc-cytf complex did not produce a converged ensemble of structures. Simulations of the dynamics were performed using the observed averaged NMR parameters as input. The results indicate a surprisingly large amplitude of mobility of Y12G/P14L Pc within the complex. It is concluded that the double mutation shifts the complex further from the well-defined toward the encounter state.
- Leiden Institute of Chemistry, Leiden University, Gorlaeus Laboratories, P.O. Box 9502, 2300 RA Leiden, The Netherlands.
Organizational Affiliation: