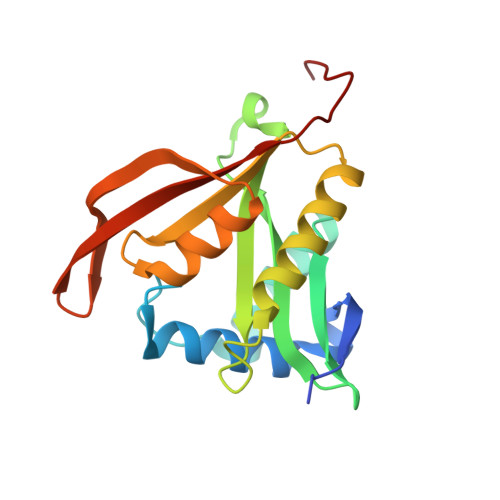
Structure and Substrate Specificity of Acetyltransferase Aciad1637 from Acinetobacter Baylyi Adp1.
Davies, A.M., Tata, R., Snape, A., Sutton, B.J., Brown, P.R.(2009) Biochimie 91: 484
- PubMed: 19135125
- DOI: https://doi.org/10.1016/j.biochi.2008.12.003
- Primary Citation of Related Structures:
2JLM - PubMed Abstract:
Gene ACIAD1637 from Acinetobacter baylyi ADP1 encodes a 182 amino acid putative antibiotic resistance protein. The structure of this protein (termed acepita) has been solved in space group P(2) to 2.35 A resolution. Acepita belongs to the GCN5-related N-acetyltransferase (GNAT) family, and contains the four sequence motifs conserved among family members. The structure of acepita is compared with that of pita, its homologue from Pseudomonas aeruginosa. Acepita has a similar substrate profile to pita and performs a similar function.
- King's College London, Randall Division of Cell and Molecular Biophysics, London, SE1 1UL, UK.
Organizational Affiliation: