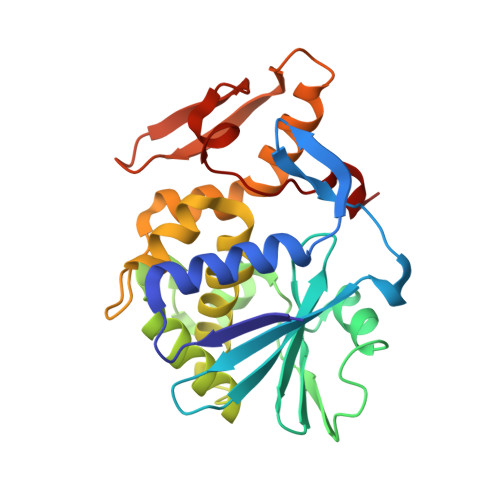
The C-Terminal Fragment of the Ribosomal P Protein Complexed to Trichosanthin Reveals the Interaction between the Ribosome-Inactivating Protein and the Ribosome.
Too, P.H., Ma, M.K., Mak, A.N., Wong, Y.T., Tung, C.K., Zhu, G., Au, S.W., Wong, K.B., Shaw, P.C.(2009) Nucleic Acids Res 37: 602
- PubMed: 19073700
- DOI: https://doi.org/10.1093/nar/gkn922
- Primary Citation of Related Structures:
2JDL, 2JJR, 2VS6 - PubMed Abstract:
Ribosome-inactivating proteins (RIPs) inhibit protein synthesis by enzymatically depurinating a specific adenine residue at the sarcin-ricin loop of the 28S rRNA, which thereby prevents the binding of elongation factors to the GTPase activation centre of the ribosome. Here, we present the 2.2 A crystal structure of trichosanthin (TCS) complexed to the peptide SDDDMGFGLFD, which corresponds to the conserved C-terminal elongation factor binding domain of the ribosomal P protein. The N-terminal region of this peptide interacts with Lys173, Arg174 and Lys177 in TCS, while the C-terminal region is inserted into a hydrophobic pocket. The interaction with the P protein contributes to the ribosome-inactivating activity of TCS. This 11-mer C-terminal P peptide can be docked with selected important plant and bacterial RIPs, indicating that a similar interaction may also occur with other RIPs.
- Department of Biochemistry, Centre for Protein Science and Crystallography and Molecular Biotechnology Programme, The Chinese University of Hong Kong, Shatin, NT, Hong Kong, China.
Organizational Affiliation: