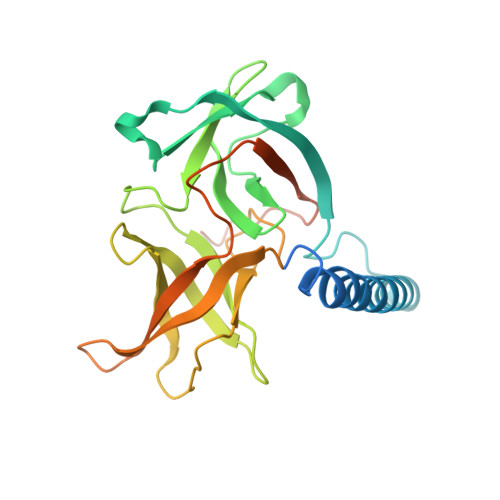
Dimeric Structure of the Cell Shape Protein Mrec and its Functional Implications.
Van Den Ent, F., Leaver, M., Bendezu, F., Errington, J., De Boer, P., Lowe, J.(2006) Mol Microbiol 62: 1631
- PubMed: 17427287
- DOI: https://doi.org/10.1111/j.1365-2958.2006.05485.x
- Primary Citation of Related Structures:
2J5U - PubMed Abstract:
The bacterial actin homologue MreB forms helical filaments in the cytoplasm of rod-shaped bacteria where it helps maintain the shape of the cell. MreB is co-transcribed with mreC that encodes a bitopic membrane protein with a major periplasmic domain. Like MreB, MreC is localized in a helical pattern and might be involved in the spatial organization of the peptidoglycan synthesis machinery. Here, we present the structure of the major, periplasmic part of MreC from Listeria monocytogenes at 2.5 A resolution. MreC forms a dimer through an intimate contact along an N-terminal alpha-helix that connects the transmembrane region with two C-terminal beta-domains. The translational relationship between the molecules enables, in principle, filament formation. One of the beta-domains shows structural similarity to the chymotrypsin family of proteins and possesses a highly conserved Thr Ser dipeptide. Unexpectedly, mutagenesis studies show that the dipeptide is dispensable for maintaining cell shape and viability in both Escherichia coil and Bacillus subtilis. Bacterial two-hybrid experiments reveal that MreC Interacts with high-molecular-weight penicillin-binding proteins (PBPs), rather than with low-molecular-weight endo- and carboxypeptidases, indicating that MreC might act as a scaffold to which the murein synthases are recruited in order to spatially organize the synthesis of new cell wall material. Deletion analyses indicate which domains of B. subtilis MreC are required for interaction with MreD as well as with the PBPs.
- MRC-LMB, Hills Road, Cambridge CB2 2QH, UK. fent@mrclmb.cam.ac.uk
Organizational Affiliation: