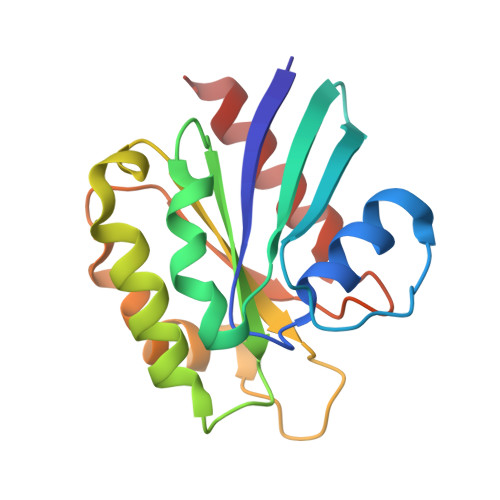
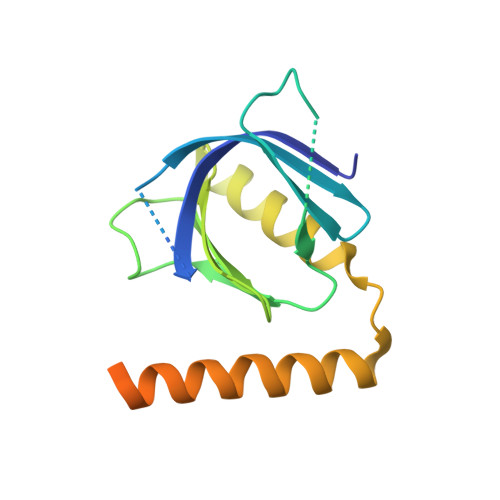
Structural Basis for Arf1-Mediated Recruitment of Arhgap21 to Golgi Membranes.
Menetrey, J., Perderiset, M., Cicolari, J., Dubois, T., El Khatib, N., El Khadali, F., Franco, M., Chavrier, P., Houdusse, A.(2007) EMBO J 26: 1953
- PubMed: 17347647
- DOI: https://doi.org/10.1038/sj.emboj.7601634
- Primary Citation of Related Structures:
2J59 - PubMed Abstract:
ARHGAP21 is a Rho family GTPase-activating protein (RhoGAP) that controls the Arp2/3 complex and F-actin dynamics at the Golgi complex by regulating the activity of the small GTPase Cdc42. ARHGAP21 is recruited to the Golgi by binding to another small GTPase, ARF1. Here, we present the crystal structure of the activated GTP-bound form of ARF1 in a complex with the Arf-binding domain (ArfBD) of ARHGAP21 at 2.1 A resolution. We show that ArfBD comprises a PH domain adjoining a C-terminal alpha helix, and that ARF1 interacts with both of these structural motifs through its switch regions and triggers structural rearrangement of the PH domain. We used site-directed mutagenesis to confirm that both the PH domain and the helical motif are essential for the binding of ArfBD to ARF1 and for its recruitment to the Golgi. Our data demonstrate that two well-known small GTPase-binding motifs, the PH domain and the alpha helical motif, can combine to create a novel mode of binding to Arfs.
- Centre de Recherche, UMR 144, Institut Curie, 26 rue d'Ulm, 75248 Paris Cedex 05, France.
Organizational Affiliation: