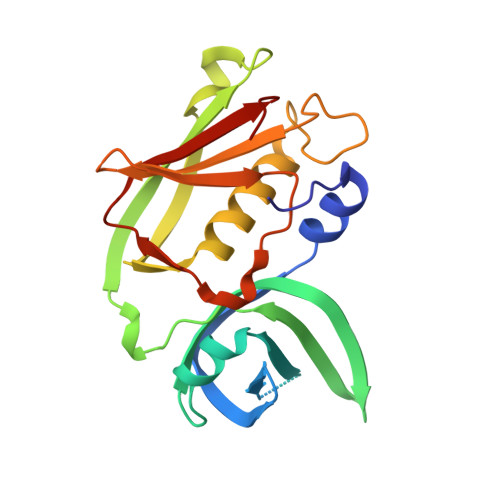
Crystal Structure of Streptococcus Dysgalactiae-Derived Mitogen Reveals a Zinc-Binding Site and Alterations in Tcr Binding.
Saarinen, S., Kato, H., Uchiyama, T., Miyoshi-Akiyama, T., Papageorgiou, A.C.(2007) J Mol Biology 373: 1089
- PubMed: 17900619
- DOI: https://doi.org/10.1016/j.jmb.2007.08.024
- Primary Citation of Related Structures:
2J4X - PubMed Abstract:
Bacterial superantigens are protein toxins with an ability to cause serious diseases in humans by activating a large number of T cells. Streptococcus dysgalactiae-derived mitogen (SDM) is a novel superantigen that is distinct from other known superantigens based on phylogenetic analysis. The X-ray structure of SDM has been determined at 1.95 A resolution. SDM shares the same characteristic fold with other superantigens, but it shows a major structural difference due to the lack of the alpha5 helix between the beta10 and beta11 strands. A bound zinc ion was identified in the structure at the C-terminal domain of the molecule. SDM appears to bind to the major histocompatibility complex class II beta-chain through the zinc-binding site, as described by mutagenesis data and structural comparisons. T-cell binding instead shows a significant difference compared to other superantigens. The mutation of Asn11 (a conserved residue that is known to be significant for T-cell-receptor binding in other superantigens) and Lys15 to Ala did not cause any decrease in the mitogenic activity of SDM. This observation and the lack of the alpha5 helix suggest alterations in T-cell-receptor binding.
- Turku Center for Biotechnology, University of Turku and Abo Akademi University, PO Box 123, Tykistökatu 6, BioCity, Turku 20521, Finland.
Organizational Affiliation: