Structure of Three Tandem Filamin Domains Reveals Auto-Inhibition of Ligand-Binding.
Lad, Y., Kiema, T.-R., Jiang, P., Pentikanen, O.T., Coles, C.H., Campbell, I.D., Calderwood, D.A., Ylanne, J.(2007) EMBO J 26: 3993
- PubMed: 17690686
- DOI: https://doi.org/10.1038/sj.emboj.7601827
- Primary Citation of Related Structures:
2J3S - PubMed Abstract:
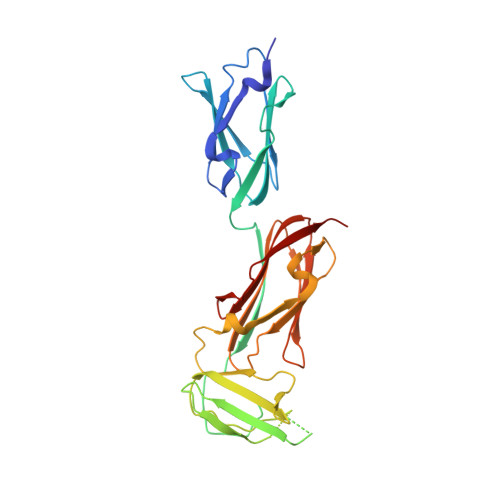
Human filamins are large actin-crosslinking proteins composed of an N-terminal actin-binding domain followed by 24 Ig-like domains (IgFLNs), which interact with numerous transmembrane receptors and cytosolic signaling proteins. Here we report the 2.5 A resolution structure of a three-domain fragment of human filamin A (IgFLNa19-21). The structure reveals an unexpected domain arrangement, with IgFLNa20 partially unfolded bringing IgFLNa21 into close proximity to IgFLNa19. Notably the N-terminus of IgFLNa20 forms a beta-strand that associates with the CD face of IgFLNa21 and occupies the binding site for integrin adhesion receptors. Disruption of this IgFLNa20-IgFLNa21 interaction enhances filamin binding to integrin beta-tails. Structural and functional analysis of other IgFLN domains suggests that auto-inhibition by adjacent IgFLN domains may be a general mechanism controlling filamin-ligand interactions. This can explain the increased integrin binding of filamin splice variants and provides a mechanism by which ligand binding might impact filamin structure.
- Department of Pharmacology and Interdepartmental Program in Vascular Biology and Transplantation, Yale University School of Medicine, New Haven, CT 06520, USA.
Organizational Affiliation: