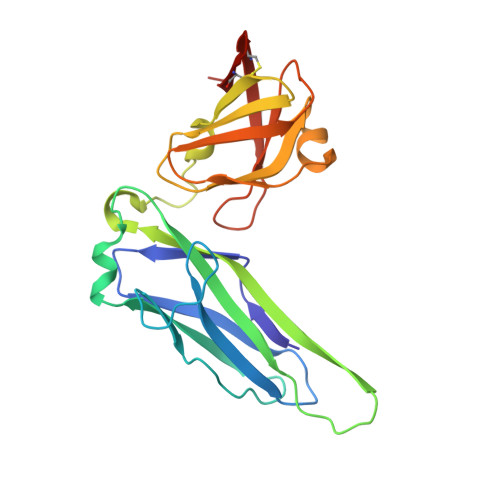
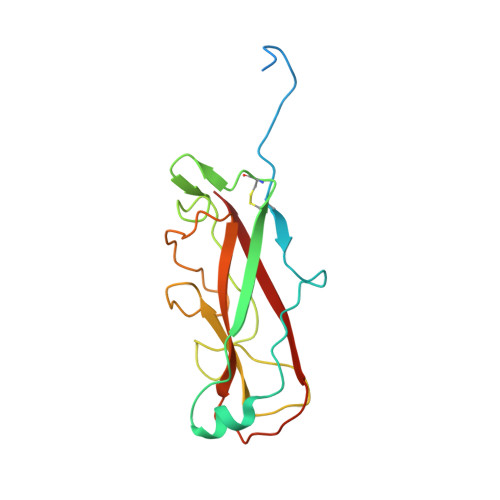
Molecular Mechanism of P Pilus Termination in Uropathogenic Escherichia Coli.
Verger, D., Miller, E., Remaut, H., Waksman, G., Hultgren, S.(2006) EMBO Rep 7: 1228
- PubMed: 17082819
- DOI: https://doi.org/10.1038/sj.embor.7400833
- Primary Citation of Related Structures:
2J2Z - PubMed Abstract:
P pili are important adhesive fibres that are assembled by the conserved chaperone-usher pathway. During pilus assembly, the subunits are incorporated into the growing fibre by the donor-strand exchange mechanism, whereby the beta-strand of the chaperone, which complements the incomplete immunoglobulin fold of each subunit, is displaced by the amino-terminal extension of an incoming subunit in a zip-in-zip-out exchange process that is initiated at the P5 pocket, an exposed hydrophobic pocket in the groove of the subunit. In vivo, termination of P pilus growth requires a specialized subunit, PapH. Here, we show that PapH is incorporated at the base of the growing pilus, where it is unable to undergo donor-strand exchange. This inability is not due to a stronger PapD-PapH interaction, but to a lack of a P5 initiator pocket in the PapH structure, suggesting that PapH terminates pilus growth because it is lacking the initiation point by which donor-strand exchange proceeds.
- Institute of Structural Molecular Biology, University College London, Birkbeck College, Malet Street, London WC1E 7HX, UK.
Organizational Affiliation: