Structural and Mutational Analysis of Human Ad37 and Canine Adenovirus 2 Fiber Heads in Complex with the D1 Domain of Coxsackie and Adenovirus Receptor.
Seiradake, E., Lortat-Jacob, H., Billet, O., Kremer, E.J., Cusack, S.(2006) J Biological Chem 281: 33704
- PubMed: 16923808
- DOI: https://doi.org/10.1074/jbc.M605316200
- Primary Citation of Related Structures:
2J12, 2J1K, 2J2J - PubMed Abstract:
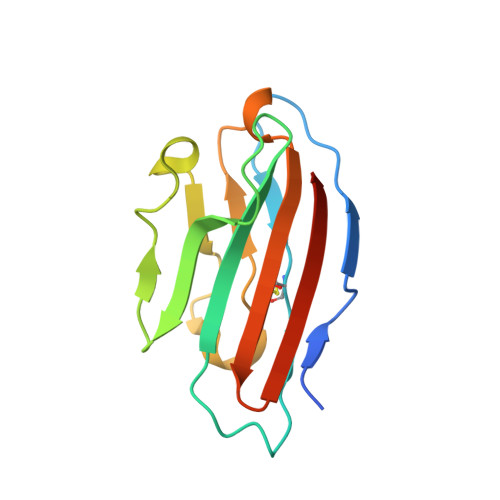
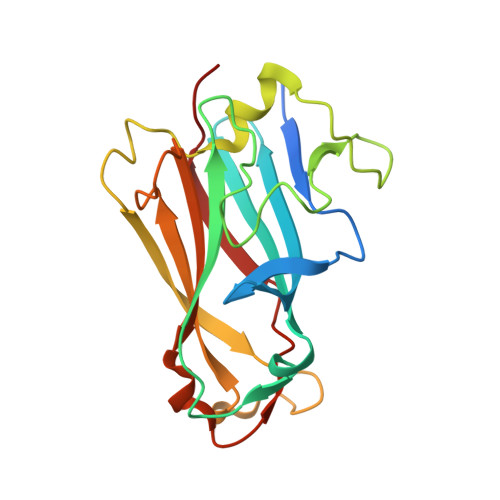
Adenovirus fibers from most serotypes bind the D1 domain of coxsackie and adenovirus receptor (CAR), although the binding residues are not strictly conserved. To understand this further, we determined the crystal structures of canine adenovirus serotype 2 (CAV-2) and the human adenovirus serotype 37 (HAd37) in complex with human CAR D1 at 2.3 and 1.5A resolution, respectively. Structure comparison with the HAd12 fiber head-CAR D1 complex showed that the overall topology of the interaction is conserved but that the interfaces differ in number and identity of interacting residues, shape complementarity, and degree of conformational adaptation. Using surface plasmon resonance, we characterized the binding affinity to CAR D1 of wild type and mutant CAV-2 and HAd37 fiber heads. We found that CAV-2 has the highest affinity but fewest direct interactions, with the reverse being true for HAd37. Moreover, we found that conserved interactions can have a minor contribution, whereas serotype-specific interactions can be essential. These results are discussed in the light of virus evolution and design of adenovirus vectors for gene transfer.
- Grenoble Outstation, European Molecular Biology Laboratory, Grenoble, France.
Organizational Affiliation: