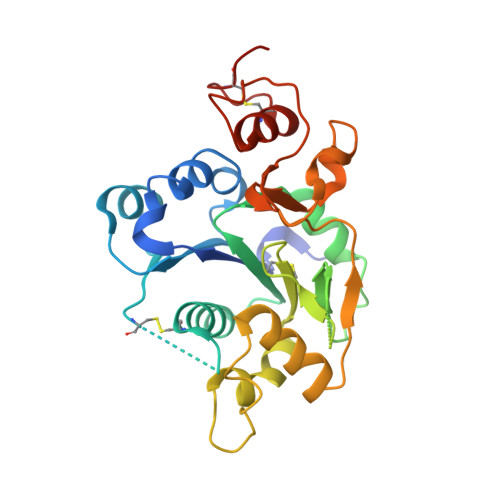
Structural Insights Into the Notch-Modifying Glycosyltransferase Fringe
Jinek, M., Chen, Y.-W., Clausen, H., Cohen, S.M., Conti, E.(2006) Nat Struct Mol Biol 13: 945
- PubMed: 16964258
- DOI: https://doi.org/10.1038/nsmb1144
- Primary Citation of Related Structures:
2J0A, 2J0B - PubMed Abstract:
Fringe proteins are beta1,3-N-acetylglucosaminyltransferases that modify Notch receptors, altering their ligand-binding specificity to regulate Notch signaling in development. We present the crystal structure of mouse Manic Fringe bound to UDP and manganese. The structure reveals amino acid residues involved in recognition of donor substrates and catalysis, and a putative binding pocket for acceptor substrates. Mutations of several invariant residues in this pocket impair Fringe activity in vivo.
- European Molecular Biology Laboratory, Meyerhofstrasse 1, D-69117 Heidelberg, Germany.
Organizational Affiliation: