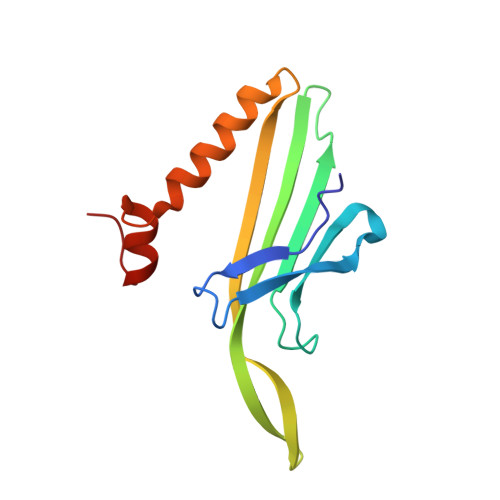
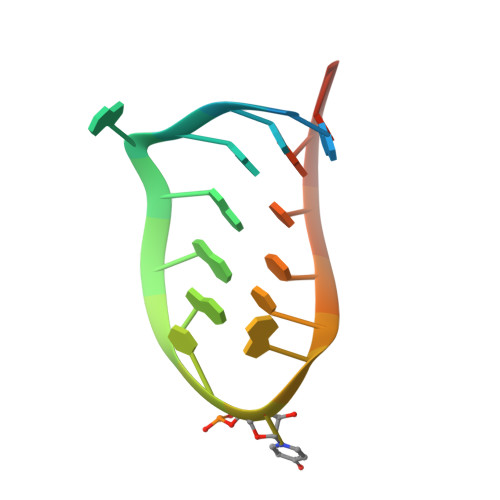
Deletion of a Single Hydrogen Bonding Atom from the MS2 RNA Operator Leads to Dramatic Rearrangements at the RNA-Coat Protein Interface
Grahn, E., Stonehouse, N.J., Adams, C.J., Fridborg, K., Beigelman, L., Matulic-Adamic, J., Warriner, S.L., Stockley, P.G., Liljas, L.(2000) Nucleic Acids Res 28: 4611
- PubMed: 11095669
- DOI: https://doi.org/10.1093/nar/28.23.4611
- Primary Citation of Related Structures:
2IZ9 - PubMed Abstract:
The MS2 coat protein binds specifically to an RNA hairpin formed within the viral genome. By soaking different RNA fragments into crystals of MS2 coat protein capsids it is possible to determine the X-ray structure of the RNA-protein complexes formed. Here we present the structure to 2.85 A resolution of a complex between a chemically modified RNA hairpin variant and the MS2 coat protein. This RNA variant has a substitution at the -5 base position, which has been shown previously to be pyrimidine-specific and is a uracil in the wild-type RNA. The modified RNA hairpin contains a pyridin-4-one base (4one) at this position that lacks the exocyclic 2-oxygen eliminating the possibility of forming a hydrogen bond to asparagine A87 in the protein. The 4one complex structure shows an unprecedented major conformational change in the loop region of the RNA, whereas there is almost no change in the conformation of the protein.
- Department of Cell and Molecular Biology, Uppsala University, Box 596, SE-751 24 Uppsala, Sweden.
Organizational Affiliation: