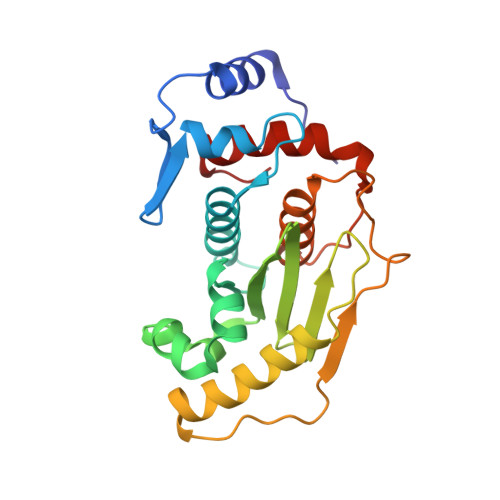
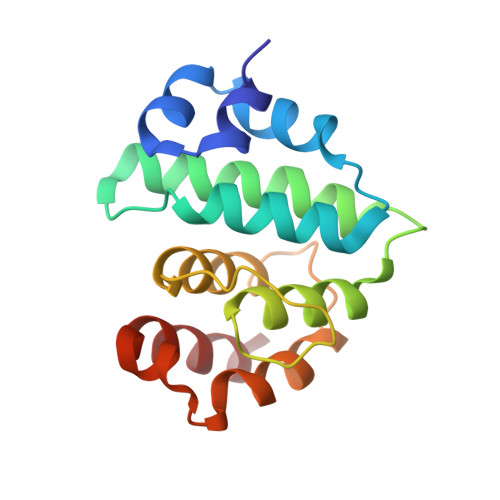
Sumo Protease Senp1 Induces Isomerization of the Scissile Peptide Bond.
Shen, L., Tatham, M.H., Dong, C., Zagorska, A., Naismith, J.H., Hay, R.T.(2006) Nat Struct Mol Biol 13: 1069
- PubMed: 17099698
- DOI: https://doi.org/10.1038/nsmb1172
- Primary Citation of Related Structures:
2IY0, 2IY1 - PubMed Abstract:
Small ubiquitin-like modifier (SUMO)-specific protease SENP1 processes SUMO-1, SUMO-2 and SUMO-3 to mature forms and deconjugates them from modified proteins. To establish the proteolytic mechanism, we determined structures of catalytically inactive SENP1 bound to SUMO-1-modified RanGAP1 and to unprocessed SUMO-1. In each case, the scissile peptide bond is kinked at a right angle to the C-terminal tail of SUMO-1 and has the cis configuration of the amide nitrogens. SENP1 preferentially processes SUMO-1 over SUMO-2, but binding thermodynamics of full-length SUMO-1 and SUMO-2 to SENP1 and K(m) values for processing are very similar. However, k(cat) values differ by 50-fold. Thus, discrimination between unprocessed SUMO-1 and SUMO-2 by SENP1 is based on a catalytic step rather than substrate binding and is likely to reflect differences in the ability of SENP1 to correctly orientate the scissile bonds in SUMO-1 and SUMO-2.
- Centre for Interdisciplinary Research, School of Life Science, University of Dundee, DD1 5EH, UK.
Organizational Affiliation: